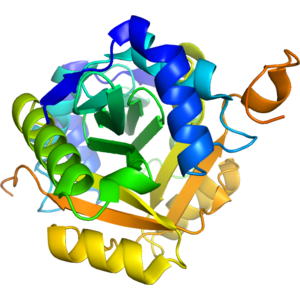
Structure of IDP92645
1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655
Edit deposit information
- CSGID target
- IDP92645
- PDB Id
- 4QUS (NCBI MMDB)
- Authors
- A.S.Halavaty,E.V.Filippova,G.Minasov,K.J.Flores,I.Dubrovska,L.Shuvalova,W.F.Anderson,Center For Structural Genomics Of Infectious Diseases (Csgid)
- Responsible person
- Andrei Halavaty
- Responsible lab
- Northwestern University
- Deposition Date
- Jul 11, 2014
- Release Date
- Jul 23, 2014
Annotation
- Description
- The 1.28 A resolution crystal structure of acyl-CoA N-acyltransferase domain (ypeA) of putative acyltransferase from Escherichia coli str. K-12 substr. MG1655 was determined by molecular replacement. There are two similar in the tertiary structure (0.8 A r.ms.d. over 139 C-alpha atoms) copies of the protein within the I4 asymmetric unit that form a dimer (5800 A2). 38 hydrogen-bonded and 333 van der Waals contacts hold the dimer together. Crystal packing demonstrates that the domain may form octamers (32700 A2) too. The acyl-CoA N-acyltransferase domain is organized into a compact core subdomain with an antiparallel 4-stranded beta-sheet that is flanked by 4 helices. Additional strand-helix-strand structural motif represents a dimerization subdomain domain. There is an extended solvent-exposed groove between the dimerization and core domain that serves as a docking site for acyl-CoA molecule co-crystallized with the acyl-CoA N-acyltransferase domain (ypeA) (see Annotation for the PDB entry 4qvt).
- Functional assignment
- Transferase
Ligands
| Ligand code | Name | Ligand type |
|---|---|---|
| 175 | 3,5-dihydro-5-methylidene-4h-imidazol-4-on | biological |
| EDO | ethylene diol | crystallization |
Structure information
Unit cell parameters
- Space Group
- I 4
- Unit Cell
-
a=90.82Å, b=90.82Å, c=84.17Å
α=90.00, β=90.00, γ=90.00 - Solvent content
- Matthews coefficient
- Resolution range
- 28.72-1.28Å (1.31-1.28Å)
- Rall(%)
- 11.1
- Rwork(%)
- 10.9 (19.0)
- Rfree(%)
- 13.9 (23.4)
- Num. observed reflections
- 87232 (6328)
- Num. Rfree reflections
- 4361 (314)
- Completeness(%)
- 99.7 (98.5)
- Num Atoms
- 2528
- Num Waters
- 537
- Num Hetatoms
- 573
- Model mean isotropic B factor
- 20.900Å2
- RMSD bond length
- 0.010Å
- RMSD bond angle
- 1.643°
- Filename uploaded
- 4QUS.pdb (uploaded on Jul 24, 2014 12:33 PM)
- Inserted
- Jul 24, 2014