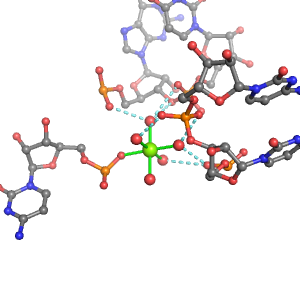
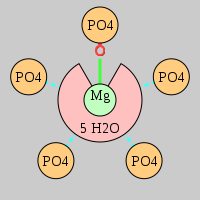
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1jj2 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03174, 07405, 08559, 09158, 09751 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
1jj2 |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
03243, 07356, 07357, 07411, 07412 |
|
C:01456, C:01483, A:01485, C:01455 |
|
G:01484, U:01457 |
|
1k73 |
A8012 |
Oph·4Pout |
G:A28(OP1) |
|
|
A3226, A3642, A7869, A9038, A9636 |
|
C:A1305, U:A1304, C:A1343, G:A1344 |
|
|
|
1k9m |
A8012 |
Oph·4Pout |
G:A28(OP1) |
|
|
A3220, A3638, A7868, A9039, A9630 |
|
C:A1305, U:A1304, C:A1343, G:A1344 |
|
|
|
1kc8 |
A8091 |
Oph·4Pout |
G:A918(OP1) |
|
|
A3321, A6482, A7851, A9095, A9544 |
|
C:A2294, C:A2391, A:A2465, U:A919 |
|
A:A2465 |
|
1kd1 |
A8012 |
Oph·4Pout |
G:A28(OP1) |
|
|
A3226, A3643, A7855, A9037, A9630 |
|
C:A1305, U:A1304, C:A1343, G:A1344 |
|
|
|
1kqs |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03155, 07340, 08560, 09152, 09735 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
1m1k |
A8012 |
Oph·4Pout |
G:A28(OP1) |
|
|
A3241, A7446, A8635, A9231, A9825 |
|
C:A1305, U:A1304, C:A1343, G:A1344 |
|
|
|
1m90 |
A8091 |
Oph·4Pout |
G:A918(OP1) |
|
|
A6020, A7382, A8612, A9068, A9848 |
|
C:A2294, C:A2391, A:A2465, G:A2293 |
|
A:A2465 |
|
1m90 |
A8035 |
Oph·4Pout |
G:A1484(OP2) |
|
|
A3244, A7339, A7340, A7394, A7395 |
|
C:A1456, C:A1483, A:A1485, C:A1455 |
|
G:A1484, U:A1457 |
|
1m90 |
A8012 |
Oph·4Pout |
G:A28(OP1) |
|
|
A3176, A7388, A8558, A9157, A9752 |
|
C:A1305, U:A1304, C:A1343, G:A1344 |
|
|
|
1n8r |
A8012 |
Oph·4Pout |
G:A28(OP1) |
|
|
A3031, A3617, A4043, A8412, A9443 |
|
C:A1343, G:A1344, C:A1305, U:A1304 |
|
|
|
1nji |
A8035 |
Oph·4Pout |
G:A1484(OP2) |
|
|
A3707, A7800, A7801, A7856, A7857 |
|
C:A1456, C:A1483, A:A1485, C:A1455 |
|
G:A1484, U:A1457 |
|
1q82 |
A8012 |
Oph·4Pout |
G:A28(OP1) |
|
|
A3161, A7329, A8559, A9151, A9738 |
|
C:A1305, U:A1304, C:A1343, G:A1344 |
|
|
|
1q86 |
A8091 |
Oph·4Pout |
G:A918(OP1) |
|
|
A5999, A7354, A8614, A9072, A9845 |
|
C:A2294, C:A2391, A:A2465, U:A919 |
|
A:A2465 |
|
1qvg |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
03221, 07189, 07190, 07243, 07244 |
|
C:01483, C:01455, C:01456, A:01485 |
|
U:01457 |
|
1qvg |
08091 |
Oph·4Pout |
G:0918(OP1) |
|
|
05909, 07232, 08616, 09065, 09830 |
|
C:02294, C:02391, A:02465, U:0919 |
|
A:02465 |
|
1s72 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03169, 07360, 08559, 09156, 09747 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
1s72 |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
03238, 07311, 07312, 07367, 07368 |
|
C:01456, C:01483, A:01485, C:01455 |
|
G:01484, U:01457 |
|
1vq4 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03534, 03953, 08131, 09359, 09956 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
1vq5 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03548, 03964, 08175, 09361, 09962 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
1vq5 |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
04030, 08126, 08127, 08182, 08183 |
|
C:01483, A:01485, C:01455, C:01456 |
|
G:01484, U:01457 |
|
1vq6 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03534, 03956, 08139, 09361, 09957 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
1vq6 |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
04024, 07983, 07984, 08146, 08147 |
|
C:01483, C:01455, A:01485, C:01456 |
|
U:01457 |
|
1vq7 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03549, 03972, 08170, 09360, 09960 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|