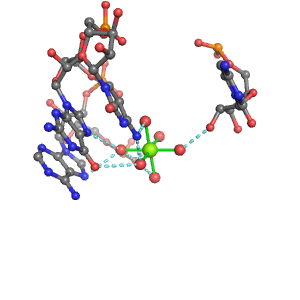
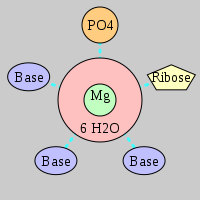
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1fuf |
A97 |
Pout·Rout·3Bout |
|
|
|
A92, A93, A96, B94, B95 |
|
U:A7 |
U:A6 |
U:B17, G:B15, G:B16 |
|
1vs5 |
A1599 |
Pout·Rout·3Bout |
|
|
|
A1872, A1873, A1874, A1875, A1876, A1877 |
|
C:A1400 |
C:A1399 |
G:A926, G:A1505, C:A1399 |
|
2i2v |
B2959 |
Pout·Rout·3Bout |
|
|
|
B3286, B3287, B3288, B3289, B3290, B3291 |
|
A:B582 |
C:B1251 |
C:B584, A:B582, G:B583 |
|
2qow |
A1546 |
Pout·Rout·3Bout |
|
|
|
A1896, A1897, A1898, A1899, A1900, A1901 |
|
G:A803 |
G:A803 |
U:A772, G:A773, G:A771 |
|
2qp1 |
B2957 |
Pout·Rout·3Bout |
|
|
|
B3773, B3774, B3775, B3776, B3777, B3778 |
|
A:B2386 |
U:B2384 |
C:B2326, U:B2387, A:B2386 |
|
2z4l |
B2983 |
Pout·Rout·3Bout |
|
|
|
B3380, B3381, B3382, B3383, B3384, B3385 |
|
A:B2386 |
U:B2384 |
C:B2326, A:B2386, U:B2387 |
|
3df1 |
A2219 |
Pout·Rout·3Bout |
|
|
|
A4220, A4221, A4222, A4223, A4224, A4225 |
|
U:A916 |
U:A13 |
U:A13, U:A916, G:A917 |
|
3i1r |
A2941 |
Pout·Rout·3Bout |
|
|
|
A3214, A3215, A3216, A3217, A3218, A3219 |
|
U:A1267 |
G:A1266 |
G:A2010, U:A2011, G:A2012 |
|
3oat |
A2982 |
Pout·Rout·3Bout |
|
|
|
A3399, A3400, A3401, A3402, A3403, A3404 |
|
C:A2055 |
A:A2054 |
U:A2034, G:A2035, A:A574 |
|
3oat |
A2940 |
Pout·Rout·3Bout |
|
|
|
A3206, A3207, A3208, A3209, A3210, A3211 |
|
U:A1267 |
G:A1266 |
G:A2010, U:A2011, G:A2012 |
|
3ofr |
A2984 |
Pout·Rout·3Bout |
|
|
|
A3409, A3410, A3411, A3412, A3413, A3414 |
|
C:A2055 |
A:A2054 |
U:A2034, G:A2035, A:A574 |
|
3ofz |
A2982 |
Pout·Rout·3Bout |
|
|
|
A3401, A3402, A3403, A3404, A3405, A3406 |
|
C:A2055 |
A:A2054 |
U:A2034, G:A2035, A:A574 |
|
3v27 |
A3573 |
Pout·Rout·3Bout |
|
|
|
A4976, A4977, A4978, A4979 |
|
U:A1955 |
G:A1954 |
G:A1950, G:A1948, G:A1949 |
|
3v27 |
A3597 |
Pout·Rout·3Bout |
|
|
|
A5056, A5057, A5058, A5059 |
|
G:A582 |
C:A1251 |
G:A582, G:A583, G:A585 |
|
4bpe |
A5001 |
Pout·Rout·3Bout |
|
|
|
A2148, A2149, A2161, A2162, A2163, A2245 |
|
G:A854 |
U:A913 |
G:A854, U:A913, A:A911 |
|
4bpn |
A5001 |
Pout·Rout·3Bout |
|
|
|
A6006, A6007, A6008, A6009, A6010, A6011 |
|
G:A854 |
U:A913 |
U:A913, G:A854, A:A911 |
|
4bpn |
A5025 |
Pout·Rout·3Bout |
|
|
|
A6433, A6434, A6435, A6436, A6437, A6438 |
|
C:A419 |
C:A416 |
G:A421, C:A416, A:A420 |
|
4bpo |
A5025 |
Pout·Rout·3Bout |
|
|
|
A2067, A2068, A2069, A2070, A2071, A2072 |
|
C:A419 |
C:A416 |
G:A421, C:A416, A:A420 |
|
4bpp |
A5001 |
Pout·Rout·3Bout |
|
|
|
A7027, A7028, A7029, A7030, A7031, A7032 |
|
G:A854 |
U:A913 |
U:A913, G:A854, A:A911 |
|
4dha |
A3742 |
Pout·Rout·3Bout |
|
|
|
A4879, A4880, A4881, A4882 |
|
U:A1955 |
G:A1954 |
G:A1948, G:A1949, G:A1950 |
|
4gar |
A3092 |
Pout·Rout·3Bout |
|
|
|
A3632, A3633, A3634, A3635 |
|
A:A1439 |
U:A1438 |
U:A1440, A:A1552, A:A1551 |
|
4gau |
A3078 |
Pout·Rout·3Bout |
|
|
|
A3572, A3573, A3574, A3575, A3576, A3577 |
|
U:A2034 |
A:A2054 |
G:A2035, A:A574, U:A2034 |
|
4nvw |
A3102 |
Pout·Rout·3Bout |
|
|
|
A4063, A4064, A4065, A4066 |
|
G:A21 |
G:A567 |
A:A573, A:A563, G:A567 |
|
4nvx |
A3294 |
Pout·Rout·3Bout |
|
|
|
A4147, A4148, A4149, A4150 |
|
C:A1333 |
U:A1313 |
U:A1335, G:A1334, U:A1316 |
|
4nw0 |
A3102 |
Pout·Rout·3Bout |
|
|
|
A4063, A4064, A4065, A4066 |
|
G:A21 |
G:A567 |
G:A567, A:A573, A:A563 |
|