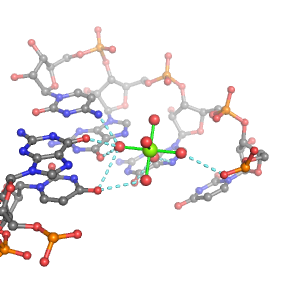
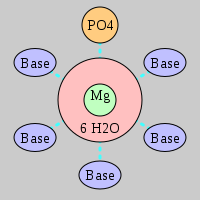
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2avy |
A1548 |
Pout·5Bout |
|
|
|
A1627, A1628, A1629, A1630, A1631, A1632 |
|
G:A645 |
|
U:A593, U:A594, U:A644, G:A645, G:A646 |
|
2qbb |
A2114 |
Pout·5Bout |
|
|
|
A2461, A2462, A2463, A2464, A2465, A2466 |
|
U:A644 |
|
U:A593, G:A592, G:A646, C:A647, G:A645 |
|
3ccr |
08028 |
Pout·5Bout |
|
|
|
03113, 03633, 03659, 03682, 07809, 09297 |
|
G:02275 |
|
G:02271, A:02274, G:02275, U:02276, U:02115 |
|
3df1 |
A2199 |
Pout·5Bout |
|
|
|
A4200, A4201, A4202, A4203, A4204 |
|
U:A1348 |
|
G:A1370, G:A1371, U:A1351, C:A1352, U:A1372 |
|
3i1o |
A1540 |
Pout·5Bout |
|
|
|
A1616, A1617, A1618, A1619, A1620, A1621 |
|
G:A645 |
|
G:A646, U:A594, G:A645, G:A592, U:A593 |
|
3ofd |
A2979 |
Pout·5Bout |
|
|
|
A3397, A3398, A3399, A3400, A3401, A3402 |
|
G:A2454 |
|
G:A2454, C:A2496, A:A2497, G:A2455, A:A2453 |
|
3ofp |
A1561 |
Pout·5Bout |
|
|
|
A1704, A1705, A1706, A1707, A1708 |
|
A:A1349 |
|
U:A1351, G:A1371, U:A1372, A:A1350, G:A1370 |
|
3v24 |
A1668 |
Pout·5Bout |
|
|
|
A1802, A1803, A1804, A1805, A1806, A1807 |
|
G:A1057 |
|
C:A1059, G:A1058, G:A1198, U:A1199, G:A1197 |
|
4dr4 |
A1666 |
Pout·5Bout |
|
|
|
A2111, A2112, A2113, A2114, A2115 |
|
A:A583 |
|
G:A584, G:A585, C:A756, U:A757, G:A758 |
|
4dr6 |
A1850 |
Pout·5Bout |
|
|
|
A2564, A2565, A2566, A2567, A2568, A2569 |
|
G:A1057 |
|
G:A1058, G:A1053, U:A1199, C:A1059, G:A1198 |
|
4dr7 |
A1827 |
Pout·5Bout |
|
|
|
A2527, A2528, A2529, A2530, A2531, A2532 |
|
G:A1057 |
|
G:A1058, C:A1059, G:A1053, G:A1198, U:A1199 |
|
4dv7 |
A1866 |
Pout·5Bout |
|
|
|
A2250, A2251, A2252, A2253, A2254, A2255 |
|
G:A266 |
|
G:A257, G:A258, U:A256, G:A266, G:A255 |
|
4gas |
A1612 |
Pout·5Bout |
|
|
|
A1754, A1755, A1756, A1757, A1758, A1759 |
|
A:A415 |
|
G:A425, G:A416, U:A426, G:A417, U:A427 |
|
4pe9 |
A1604 |
Pout·5Bout |
|
|
|
A1716, A1717, A1718, A1719, A1720, A1721 |
|
A:A415 |
|
G:A416, U:A427, G:A417, C:A418, G:A425 |
|
4too |
A3066 |
Pout·5Bout |
|
|
|
A3495, A3496, A3497, A3498, A3499, A3500 |
|
C:A2073 |
|
G:A2242, U:A2243, U:A2074, G:A2238, A:A2241 |
|
4tov |
A3085 |
Pout·5Bout |
|
|
|
A3584, A3585, A3586, A3587, A3588, A3589 |
|
A:A933 |
|
U:A934, G:A841, U:A842, G:A843, A:A936 |
|
4tox |
A3072 |
Pout·5Bout |
|
|
|
A3522, A3523, A3524, A3525, A3526, A3527 |
|
A:A2453 |
|
A:A2453, G:A2454, A:A2497, G:A2455, C:A2496 |
|
4tp3 |
A3071 |
Pout·5Bout |
|
|
|
A3521, A3522, A3523, A3524, A3525, A3526 |
|
A:A2453 |
|
A:A2497, A:A2453, G:A2454, G:A2455, C:A2496 |
|