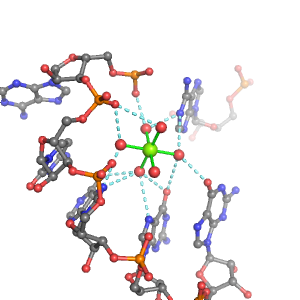
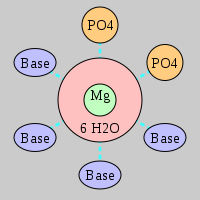
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3q3z |
V5 |
2Pout·4Bout |
|
|
|
V100, V104, V107, V108, V109, V110 |
|
C:V72, G:V73 |
|
G:V12, A:V74, A:V13, G:V73 |
|
4a17 |
063 |
2Pout·4Bout |
|
|
|
05603, 05604, 05605, 05606, 05607, 05608 |
|
A:257, A:256 |
|
U:258, G:259, G:249, C:250 |
|
4a1a |
063 |
2Pout·4Bout |
|
|
|
05491, 05492, 05493, 05494, 05495, 05496 |
|
A:257, A:256 |
|
U:258, G:249, C:250, G:259 |
|
4gau |
B201 |
2Pout·4Bout |
|
|
|
B301, B302, B303, B304 |
|
A:B57, A:B58 |
|
U:B22, C:B60, G:B23, A:B59 |
|
4a1a |
0325 |
2Pout·4Bout |
|
|
|
07715, 07716, 07717, 07718, 07719, 07720 |
|
G:365, A:364 |
|
U:3109, G:3108, G:365, G:366 |
|
3or9 |
A1537 |
2Pout·4Bout |
|
|
|
A1593, A1594, A1595, A1596, A1597, A1598 |
|
A:A415, G:A416 |
|
G:A417, C:A418, G:A416, U:A426 |
|
3ofx |
A1537 |
2Pout·4Bout |
|
|
|
A1592, A1593, A1594, A1595, A1596, A1597 |
|
A:A415, G:A416 |
|
G:A417, C:A418, G:A425, G:A416 |
|
2qbh |
A1548 |
2Pout·4Bout |
|
|
|
A1629, A1630, A1631, A1632, A1633, A1634 |
|
G:A645, U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
2qou |
A1548 |
2Pout·4Bout |
|
|
|
A1922, A1923, A1924, A1925, A1926, A1927 |
|
G:A645, U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
3i21 |
A1553 |
2Pout·4Bout |
|
|
|
A1681, A1682, A1683, A1684, A1685, A1686 |
|
C:A797, A:A781 |
|
U:A798, G:A799, G:A800, C:A797 |
|
4tp8 |
A1604 |
2Pout·4Bout |
|
|
|
A1715, A1716, A1717, A1718, A1719, A1720 |
|
G:A416, A:A415 |
|
G:A416, G:A417, U:A426, G:A425 |
|
4gd1 |
A1604 |
2Pout·4Bout |
|
|
|
A1717, A1718, A1719, A1720, A1721, A1722 |
|
A:A415, G:A416 |
|
G:A416, G:A417, U:A426, G:A425 |
|
4tol |
A1604 |
2Pout·4Bout |
|
|
|
A1716, A1717, A1718, A1719, A1720, A1721 |
|
A:A415, G:A416 |
|
G:A416, G:A417, G:A425, U:A426 |
|
4tp0 |
A1604 |
2Pout·4Bout |
|
|
|
A1716, A1717, A1718, A1719, A1720, A1721 |
|
A:A415, G:A416 |
|
G:A417, C:A418, U:A426, G:A416 |
|
4tp2 |
A1612 |
2Pout·4Bout |
|
|
|
A1752, A1753, A1754, A1755, A1756, A1757 |
|
G:A416, A:A415 |
|
G:A417, G:A425, U:A426, G:A416 |
|
4pea |
A1612 |
2Pout·4Bout |
|
|
|
A1750, A1751, A1752, A1753, A1754, A1755 |
|
G:A416, A:A415 |
|
G:A417, G:A425, U:A426, G:A416 |
|
4tpa |
A1622 |
2Pout·4Bout |
|
|
|
A1801, A1802, A1803, A1804, A1805, A1806 |
|
G:A803, U:A804 |
|
G:A771, U:A772, A:A807, G:A773 |
|
4tp2 |
A1622 |
2Pout·4Bout |
|
|
|
A1801, A1802, A1803, A1804, A1805, A1806 |
|
G:A803, U:A804 |
|
G:A771, U:A772, A:A807, G:A773 |
|
4ji1 |
A1756 |
2Pout·4Bout |
|
|
|
A2225, A2226, A2227, A2228, A2229, A2230 |
|
A:A431, G:A407 |
|
G:A409, G:A410, A:A408, G:A407 |
|
4dv0 |
A1810 |
2Pout·4Bout |
|
|
|
A2214, A2215, A2216, A2217, A2218 |
|
A:A759, G:A760 |
|
U:A580, G:A581, G:A579, G:A761 |
|
4dr1 |
A1814 |
2Pout·4Bout |
|
|
|
A2126, A2127, A2128, A2129 |
|
U:A264, G:A265 |
|
G:A190, G:A260, U:A261, G:A259 |
|
4dv6 |
A1838 |
2Pout·4Bout |
|
|
|
A2298, A2299, A2300, A2301, A2302, A2303 |
|
A:A759, G:A760 |
|
G:A579, U:A580, G:A761, C:A762 |
|
4ji0 |
A1858 |
2Pout·4Bout |
|
|
|
A2155, A2156, A2157, A2158, A2159, A2160 |
|
A:A609, G:A610 |
|
C:A612, G:A627, G:A628, U:A626 |
|
2qb9 |
A2031 |
2Pout·4Bout |
|
|
|
A2384, A2385, A2386, A2387, A2388, A2389 |
|
G:A645, U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
2qal |
A2031 |
2Pout·4Bout |
|
|
|
A2032, A2033, A2034, A2035, A2036, A2037 |
|
G:A645, U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|