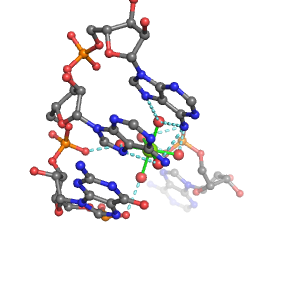
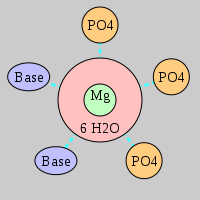
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1dfu |
N504 |
3Pout·2Bout |
|
|
|
M536, N507, N527, N528, N530, N568 |
|
A:M99, G:N75, U:N74 |
|
G:N76, G:N75 |
|
1l2x |
A38 |
3Pout·2Bout |
|
|
|
A39, A40, A41, A42, A43, A44 |
|
C:A26, G:A27, G:A28 |
|
G:A27, G:A28 |
|
1vs6 |
B2931 |
3Pout·2Bout |
|
|
|
B3143, B3144, B3145, B3146, B3147, B3148 |
|
A:B1937, A:B1936, G:B1959 |
|
G:B1959, A:B1960 |
|
1vs6 |
B2953 |
3Pout·2Bout |
|
|
|
B3245, B3246, B3247, B3248, B3249, B3250 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
1vs7 |
A1602 |
3Pout·2Bout |
|
|
|
A1886, A1887, A1888, A1889, A1890, A1891 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
1vs8 |
B2985 |
3Pout·2Bout |
|
|
|
B3405, B3406, B3407, B3408, B3409, B3410 |
|
C:B2050, A:B2051, G:B2578 |
|
A:B2051, A:B2614 |
|
1vt2 |
A2968 |
3Pout·2Bout |
|
|
|
A3343, A3344, A3345, A3346, A3347, A3348 |
|
A:A2051, G:A2578, C:A2050 |
|
A:A2051, A:A2614 |
|
2aw4 |
B2953 |
3Pout·2Bout |
|
|
|
B3241, B3242, B3243, B3244, B3245, L345 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
2aw7 |
A1601 |
3Pout·2Bout |
|
|
|
A1888, A1889, A1890, A1891, A1892, A1893 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
2awb |
B2907 |
3Pout·2Bout |
|
|
|
B3026, B3027, B3028, B3029, B3030, B3031 |
|
G:B831, U:B568, C:B944 |
|
A:B945, A:B2448 |
|
2awb |
B2986 |
3Pout·2Bout |
|
|
|
B3406, B3407, B3408, B3409, B3410, B3411 |
|
C:B2050, A:B2051, G:B2578 |
|
A:B2051, A:B2614 |
|
2i2t |
B2953 |
3Pout·2Bout |
|
|
|
B3252, B3253, B3254, B3255, B3256, L455 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
2i2u |
A1596 |
3Pout·2Bout |
|
|
|
A1856, A1857, A1858, A1859, X267, X270 |
|
G:A1401, U:X2, G:X3 |
|
C:A1399, G:A1401 |
|
2i2u |
A1574 |
3Pout·2Bout |
|
|
|
A1745, A1746, A1747, A1748, A1749, A1750 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
2i2v |
B2907 |
3Pout·2Bout |
|
|
|
B3035, B3036, B3037, B3038, B3039, B3040 |
|
C:B944, U:B568, G:B831 |
|
A:B945, A:B2448 |
|
2i2v |
B2909 |
3Pout·2Bout |
|
|
|
B3045, B3046, B3047, B3048, B3049, B3050 |
|
A:B1936, A:B1937, G:B1959 |
|
G:B1959, A:B1960 |
|
2qal |
A2044 |
3Pout·2Bout |
|
|
|
A2045, A2046, A2047, A2048, A2049, A2050 |
|
A:A315, U:A317, C:A316 |
|
G:A319, G:A318 |
|
2qam |
B3282 |
3Pout·2Bout |
|
|
|
B3840, B3841, B3842, B3843, B3844, B3845 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
2qam |
B3157 |
3Pout·2Bout |
|
|
|
B3738, B3739, B3740, B3741, B3742, B3743 |
|
A:B1937, A:B1936, G:B1959 |
|
A:B1960, G:B1959 |
|
2qao |
B3014 |
3Pout·2Bout |
|
|
|
B3630, B3631, B3632, B3633, B3634, L145 |
|
U:B568, G:B831, C:B944 |
|
A:B945, A:B2448 |
|
2qba |
B3282 |
3Pout·2Bout |
|
|
|
B3842, B3843, B3844, B3845, B3846, B3847 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
2qbc |
B3014 |
3Pout·2Bout |
|
|
|
B3631, B3632, B3633, B3634, B3635, L145 |
|
U:B568, G:B831, C:B944 |
|
A:B945, A:B2448 |
|
2qbd |
A2328 |
3Pout·2Bout |
|
|
|
A2329, A2330, A2331, A2332, A2333, A2334 |
|
A:A918, A:A1394, G:A917 |
|
A:A918, A:A919 |
|
2qbd |
A2164 |
3Pout·2Bout |
|
|
|
A2165, A2166, A2167, A2168, A2169 |
|
A:A768, U:A804, A:A767 |
|
A:A768, G:A769 |
|
2qbf |
A1600 |
3Pout·2Bout |
|
|
|
A1886, A1887, A1888, A1889, A1890, A1891 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|