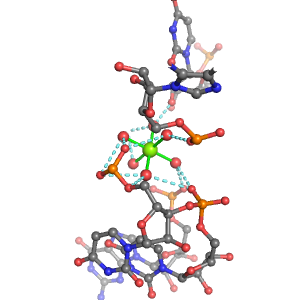
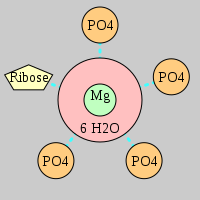
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1hr2 |
A14 |
4Pout·Rout |
|
|
|
A347, A348, A349, A350, A351, A352 |
|
A:A113, U:A202, C:A128, C:A203 |
C:A127 |
|
|
1vs8 |
B2966 |
4Pout·Rout |
|
|
|
B3315, B3316, B3317, B3318, B3319, B3320 |
|
G:B559, A:B1008, G:B2038, C:B560 |
C:B1007 |
|
|
1vs8 |
B2935 |
4Pout·Rout |
|
|
|
B3165, B3166, B3167, B3168, B3169, B3170 |
|
U:B2017, G:B579, U:B2016, G:B578 |
U:B1255 |
|
|
2aw4 |
B2955 |
4Pout·Rout |
|
|
|
B3251, B3252, B3253, B3254, B3255, E206 |
|
G:B579, U:B2017, U:B2016, A:B2058 |
U:B1255 |
|
|
2awb |
B2936 |
4Pout·Rout |
|
|
|
B3169, B3170, B3171, B3172, B3173, B3174 |
|
U:B2017, G:B579, U:B2016, G:B578 |
U:B1255 |
|
|
2qam |
B3295 |
4Pout·Rout |
|
|
|
B3851, B3852, B3853, B3854, B3855, B3856 |
|
G:B578, U:B2016, G:B579, U:B2017 |
U:B1255 |
|
|
2qao |
B3189 |
4Pout·Rout |
|
|
|
B3774, B3775, B3776, B3777, B3778, B3779 |
|
G:B579, U:B2017, U:B2016, G:B578 |
U:B1255 |
|
|
2qba |
B3295 |
4Pout·Rout |
|
|
|
B3853, B3854, B3855, B3856, B3857, B3858 |
|
G:B578, U:B2016, G:B579, U:B2017 |
U:B1255 |
|
|
2qbc |
B3189 |
4Pout·Rout |
|
|
|
B3774, B3775, B3776, B3777, B3778, B3779 |
|
G:B579, U:B2017, U:B2016, G:B578 |
U:B1255 |
|
|
2qbe |
B3295 |
4Pout·Rout |
|
|
|
B3851, B3852, B3853, B3854, B3855, B3856 |
|
G:B578, U:B2016, G:B579, U:B2017 |
U:B1255 |
|
|
2qbf |
A1596 |
4Pout·Rout |
|
|
|
A1868, A1869, A1870, A1871, A1872 |
|
C:A536, G:A537, A:A523, C:A503 |
C:A522 |
|
|
2qbg |
B2936 |
4Pout·Rout |
|
|
|
B3173, B3174, B3175, B3176, B3177, B3178 |
|
U:B2017, G:B578, G:B579, U:B2016 |
U:B1255 |
|
|
2qbg |
B2968 |
4Pout·Rout |
|
|
|
B3326, B3327, B3328, B3329, B3330, B3331 |
|
G:B559, C:B560, G:B2038, A:B1008 |
C:B1007 |
|
|
2qbi |
B2955 |
4Pout·Rout |
|
|
|
B3252, B3253, B3254, B3255, B3256, B3257 |
|
G:B578, G:B579, U:B2016, U:B2017 |
U:B1255 |
|
|
2qov |
B2955 |
4Pout·Rout |
|
|
|
B3255, B3256, B3257, B3258, B3259, B3260 |
|
G:B578, U:B2016, G:B579, U:B2017 |
U:B1255 |
|
|
2qox |
B2936 |
4Pout·Rout |
|
|
|
B3170, B3171, B3172, B3173, B3174, B3175 |
|
G:B578, U:B2017, U:B2016, G:B579 |
U:B1255 |
|
|
2qoz |
B2955 |
4Pout·Rout |
|
|
|
B3746, B3747, B3748, B3749, B3750, B3751 |
|
G:B578, G:B579, U:B2016, U:B2017 |
U:B1255 |
|
|
2qp1 |
B2936 |
4Pout·Rout |
|
|
|
B3676, B3677, B3678, B3679, B3680, B3681 |
|
G:B579, U:B2017, U:B2016, G:B578 |
U:B1255 |
|
|
2z4l |
B2909 |
4Pout·Rout |
|
|
|
B3033, B3034, B3035, B3036, B3037, B3038 |
|
C:B560, A:B1008, G:B559, G:B2038 |
C:B1007 |
|
|
2z4n |
B2937 |
4Pout·Rout |
|
|
|
B3172, B3173, B3174, B3175, B3176, B3177 |
|
G:B579, U:B2017, U:B2016, G:B578 |
U:B1255 |
|
|
2z4n |
B2969 |
4Pout·Rout |
|
|
|
B3324, B3325, B3326, B3327, B3328, B3329 |
|
C:B560, A:B1008, G:B2038, G:B559 |
C:B1007 |
|
|
3df4 |
B3189 |
4Pout·Rout |
|
|
|
B4190, B4191, B4192, B4193, B4194, B4195 |
|
G:B579, U:B2017, G:B578, U:B2016 |
U:B1255 |
|
|
3df4 |
B3375 |
4Pout·Rout |
|
|
|
B4376, B4377, B4378, B4379, B4380, B4381 |
|
G:B559, C:B560, G:B2038, A:B1008 |
C:B1007 |
|
|
3i1p |
A2998 |
4Pout·Rout |
|
|
|
A3481, A3482, A3483, A3484, A3485, A3486 |
|
U:A1636, G:A1300, G:A1299, A:A1637 |
A:A1635 |
|
|
3i1t |
A2914 |
4Pout·Rout |
|
|
|
A3089, A3090, A3091, A3092, A3093, A3094 |
|
C:A560, A:A1008, G:A559, G:A2038 |
C:A1007 |
|
|