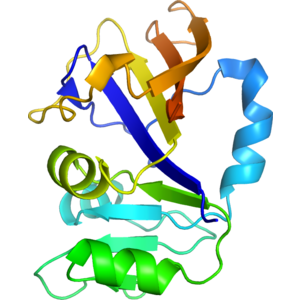
Structure of IDP90544
The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A
Edit deposit information
- CSGID target
- IDP90544
- PDB Id
- 5FDA (NCBI MMDB)
- Authors
- 'C.Chang,N.Maltseva,Y.Kim,M.Makowska-Grzyska,R.Mulligan,L.Papazisi,W.F.Anderson,A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)'
- Responsible person
- Natalia Maltseva
- Responsible lab
- Argonne National Laboratory
- Deposition Date
- Dec 15, 2015
- Release Date
- Dec 30, 2015
Annotation
- Description
- Dihydrofolate reductase is a housekeeping enzyme which is responsible for maintaining the amount of tetrahydrofolate in the cell. It catalyzes the NADPH-linked reduction of 7,8-dihydrofolate to 5,6,7,8- tetrahydrofolate. Tetrahydrofolate is the cofactor used in synthesis of several important metabolites like thymidylate, a building block of DNA. Crystal Structure of Dihydrofolate Reductase from Yersinia pestis was solved at 1.55A resolution. Protein has single domain composed of 4 alpha helices and 8 beta-sheets.
- Functional assignment
Ligands
| Ligand code | Name | Ligand type |
|---|---|---|
| CL | chloride | crystallization |
| 175 | 3,5-dihydro-5-methylidene-4h-imidazol-4-on |
Structure information
Unit cell parameters
- Space Group
- C 2 2 21
- Unit Cell
-
a=60.27Å, b=79.20Å, c=64.32Å
α=90.00, β=90.00, γ=90.00 - Solvent content
- Matthews coefficient
- Resolution range
- 47.96-1.55Å (1.62-1.55Å)
- Rall(%)
- 15.9
- Rwork(%)
- 15.7 (19.5)
- Rfree(%)
- 19.4 (27.5)
- Num. observed reflections
- 23372 (2307)
- Num. Rfree reflections
- 1168 (120)
- Completeness(%)
- 97.8 (83.0)
- Num Atoms
- 1255
- Num Waters
- 187
- Num Hetatoms
- 262
- Model mean isotropic B factor
- 19.050Å2
- RMSD bond length
- 0.009Å
- RMSD bond angle
- 0.973°
- RMSD dihedral angle
- 18.538°
- Filename uploaded
- D_1000216398_model-annotate_P1.pdb (uploaded on Jan 04, 2016 4:16 PM)
- Inserted
- Jan 04, 2016