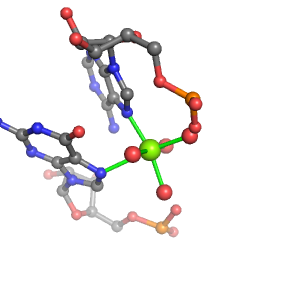
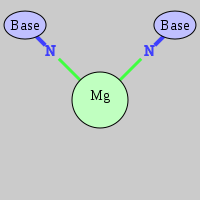
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3ofo |
A1557 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1695, A1696, A1697, A1698 |
|
G:A858, G:A869, C:A868 |
|
|
|
3ofp |
A1559 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1694, A1695, A1696, A1697 |
|
G:A858, G:A869, C:A868 |
|
|
|
3ofq |
A2925 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A3140, A3141, A3142, A3143 |
|
A:A761, G:A733, G:A760, C:A732 |
|
G:A760 |
|
3ofq |
A2950 |
2Nb |
|
|
G:A1358(N7), G:A1371(N7) |
A3255, A3256, A3257, A3258 |
|
G:A1358, C:A1357, G:A1371 |
|
|
|
3ofr |
A2923 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A3130, A3131, A3132, A3133 |
|
A:A761, G:A733, G:A760 |
C:A732 |
|
|
3ofr |
A2948 |
2Nb |
|
|
G:A1358(N7), G:A1371(N7) |
A3243, A3244, A3245, A3246 |
|
G:A1371 |
|
|
|
3ofx |
A1558 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1698, A1699, A1700, A1701 |
|
G:A858, G:A869, C:A857, C:A868 |
|
|
|
3ofy |
A1559 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1697, A1698, A1699, A1700 |
|
G:A858, G:A869, C:A868 |
|
|
|
3ofz |
A2946 |
2Nb |
|
|
G:A1358(N7), G:A1371(N7) |
A3236, A3237, A3238, A3239 |
|
G:A1358, G:A1371, C:A1357 |
|
|
|
3og0 |
A2948 |
2Nb |
|
|
G:A1358(N7), G:A1371(N7) |
A3252, A3253, A3254, A3255 |
|
G:A1358, C:A1357, G:A1371 |
|
|
|
3og0 |
A2923 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A3139, A3140, A3141, A3142 |
|
A:A761, G:A733, G:A760, C:A732 |
|
|
|
3or9 |
A1559 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1699, A1700, A1701, A1702 |
|
G:A858, G:A869, C:A868 |
|
|
|
3r8t |
A2947 |
2Nb |
|
|
G:A1358(N7), G:A1371(N7) |
A3275, A3276, A3277, A3278 |
|
G:A1371, G:A1358, C:A1357 |
|
|
|
3v23 |
A3302 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A3894, A3895, A3896, A3897 |
|
C:A732, G:A733, G:A760, A:A761 |
|
|
|
3v27 |
A3312 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A3869, A3870, A3871, A3872 |
|
C:A732, G:A733, G:A760, A:A761 |
|
|
|
3v29 |
A3343 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A3788, A3789, A3790, A3791 |
|
A:A761, G:A733, C:A732 |
|
|
|
3v2d |
A3130 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A4046, A4047, A4048, A4049 |
|
C:A732, G:A733, G:A760, A:A761 |
|
|
|
3v2f |
A3166 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A3810, A3811, A3812, A3813 |
|
G:A760, A:A761, G:A733, C:A732 |
|
|
|
4dha |
A3815 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A5142, A5143, A5144 |
|
C:A732, A:A761 |
|
|
|
4dhc |
A3384 |
2Nb |
|
|
G:A733(N7), A:A761(N7) |
A3815, A3816, A3817, A3818 |
|
G:A760, A:A761, G:A733 |
C:A732 |
|
|
4gaq |
A1625 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1817, A1818, A1819, A1820 |
|
G:A858, G:A869, C:A857 |
|
|
|
4kj0 |
A1625 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1810, A1811 |
|
G:A858, C:A868, G:A869 |
|
|
|
4kj8 |
A1625 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1813, A1814, A1816 |
|
G:A858, C:A868, G:A869 |
|
|
|
4kja |
A1624 |
2Nb |
|
|
G:A858(N7), G:A869(N7) |
A1817, A1819 |
|
G:A869 |
|
|
|
4nvv |
A3316 |
2Nb |
|
|
G:A780(N7), A:A808(N7) |
A4100, A4101, A4102, A4103 |
|
C:A779, G:A780, G:A807, A:A808 |
|
|
|