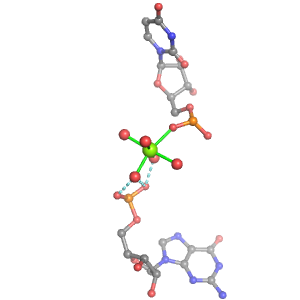
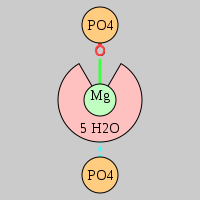
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2otl |
08092 |
Oph·Pout |
C:01844(OP2) |
|
|
03656, 03671, 07882, 09387 |
|
A:01843 |
|
A:01845 |
|
1k9m |
A8084 |
Oph·Pout |
A:A1742(OP2) |
|
|
A3626, A7858, A9249, A9370, A9741 |
|
G:A2033 |
|
G:A1744, G:A1745, A:A1742, G:A1743, G:A2033 |
|
1yij |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03644, 07818, 09250, 09377, 09755 |
|
G:02033 |
|
G:01744, G:01745, A:01742, G:01743, G:02033 |
|
1s72 |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03156, 07350, 08769, 08893, 09274 |
|
G:02033 |
|
G:01744, G:01745, A:01742, G:01743, G:02033 |
|
2otj |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03654, 07841, 09255, 09382, 09763 |
|
G:02033 |
|
G:01744, G:01745, A:01742, G:01743, G:02033 |
|
1m1k |
A8084 |
Oph·Pout |
A:A1742(OP2) |
|
|
A3229, A7436, A8841, A8964, A9347 |
|
G:A2033 |
|
G:A1744, G:A1745, A:A1742, G:A1743, G:A2033 |
|
1yi2 |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03632, 07805, 09249, 09376, 09754 |
|
G:02033 |
|
G:01744, G:01745, A:01742, G:01743, G:02033 |
|
3cc2 |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03188, 07408, 08778, 08906, 09290 |
|
G:02033 |
|
G:01744, G:01745, A:01742, G:01743, G:02033 |
|
1vq6 |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03068, 03943, 08129, 09575, 09696 |
|
G:02033 |
|
G:02033, G:01744, G:01745, A:01742, G:01743 |
|
1q82 |
A8084 |
Oph·Pout |
A:A1742(OP2) |
|
|
A3147, A7319, A8768, A8892, A9261 |
|
G:A2033 |
|
G:A1744, G:A1745, A:A1742, G:A1743, G:A2033 |
|
1vq5 |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03074, 03951, 08165, 09573, 09699 |
|
G:02033 |
|
G:02033, G:01744, G:01745, A:01742, G:01743 |
|
1vql |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03365, 04227, 08374 |
|
G:02033 |
|
G:02033, G:01743, G:01744, G:01745, A:01742 |
|
1k8a |
A8084 |
Oph·Pout |
A:A1742(OP2) |
|
|
A3225, A7397, A8844, A8963, A9342 |
|
G:A2033 |
|
G:A1744, G:A1745, A:A1742, G:A1743, G:A2033 |
|
1qvf |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03151, 08772, 08895, 09273 |
|
G:02033 |
|
G:01744, G:01745, G:01743, G:02033 |
|
1k73 |
A8084 |
Oph·Pout |
A:A1742(OP2) |
|
|
A3629, A7859, A9248, A9374, A9750 |
|
G:A2033 |
|
G:A1744, G:A1745, A:A1742, G:A1743, G:A2033 |
|
1vqm |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03367, 04234, 08375 |
|
G:02033 |
|
G:02033, G:01743, G:01744, G:01745, A:01742 |
|
3cxc |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03236, 03361, 03727, 04583, 08740 |
|
G:02033 |
|
G:01743, G:01744, G:01745, G:02033, A:01742 |
|
2otl |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03646, 07828, 09250, 09375, 09757 |
|
G:02033 |
|
G:01744, G:01745, A:01742, G:01743, G:02033 |
|
1vq7 |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03073, 03959, 08160, 09577, 09699 |
|
G:02033 |
|
G:02033, G:01744, G:01745, A:01742, G:01743 |
|
1yj9 |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03449, 07628, 09070, 09193, 09569 |
|
G:02033 |
|
G:01744, G:01745, A:01742, G:01743, G:02033 |
|
1vq4 |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03067, 03940, 08121, 09569, 09694 |
|
G:02033 |
|
G:02033, G:01744, G:01745, A:01742, G:01743 |
|
2qex |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03459, 07680, 09074, 09198, 09582 |
|
G:02033 |
|
G:01744, G:01745, A:01742, G:01743, G:02033 |
|
1vqp |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03376, 04246, 08395 |
|
G:02033 |
|
G:02033, G:01743, G:01744, G:01745, A:01742 |
|
1vqn |
08084 |
Oph·Pout |
A:01742(OP2) |
|
|
03363, 04230, 08367 |
|
G:02033 |
|
G:02033, G:01743, G:01744, G:01745, A:01742 |
|
1n8r |
A8084 |
Oph·Pout |
A:A1742(OP2) |
|
|
A3146, A4030, A8402, A9647, A9768 |
|
G:A2033 |
|
G:A2033, G:A1744, G:A1745, A:A1742, G:A1743 |
|