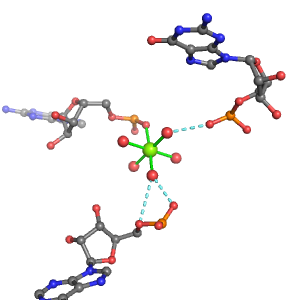
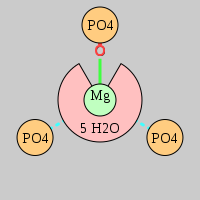
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1yi2 |
08052 |
Oph·2Pout |
U:01125(OP1) |
|
|
03621, 07849, 98590, 98616 |
|
C:01126, G:992 |
|
C:01129, G:990 |
|
1yi2 |
08019 |
Oph·2Pout |
A:02011(OP2) |
|
|
04146, 07211, 07742, 07743, 07744 |
|
A:01941, G:01891 |
U:01890 |
G:01891, C:01892 |
|
1yi2 |
08083 |
Oph·2Pout |
A:01754(OP2) |
|
|
03704, 04147, 06340, 07804, 07857 |
|
C:01753, A:01755 |
|
A:01755, A:01754 |
|
1yi2 |
08068 |
Oph·2Pout |
A:02568(OP2) |
|
|
03749, 04811, 06208, 07851, 09736 |
|
A:02569, G:02567 |
|
A:02569, A:02568 |
|
1yi2 |
08020 |
Oph·2Pout |
C:01830(OP1) |
|
|
07726, 07745, 09121, 09124, 09195 |
|
U:01831, A:01842 |
G:01828 |
G:01828, G:01832 |
|
1yij |
08068 |
Oph·2Pout |
A:02568(OP2) |
|
|
03761, 04834, 06222, 07864, 09737 |
|
A:02569, G:02567 |
|
A:02569, A:02568 |
|
1yij |
08020 |
Oph·2Pout |
C:01830(OP1) |
|
|
07738, 07757, 09123, 09126, 09198 |
|
U:01831, A:01842 |
G:01828 |
G:01828, G:01832 |
|
1yij |
08019 |
Oph·2Pout |
A:02011(OP2) |
|
|
04166, 07225, 07754, 07755, 07756 |
|
A:01941, G:01891 |
U:01890 |
G:01891, C:01892 |
|
1yij |
08083 |
Oph·2Pout |
A:01754(OP2) |
|
|
03716, 04167, 06354, 07817, 07869 |
|
C:01753, A:01755 |
|
A:01755, A:01754 |
|
1yj9 |
08020 |
Oph·2Pout |
C:01830(OP1) |
|
|
07551, 07570, 08942, 08945, 09018 |
|
U:01831, A:01842 |
G:01828 |
G:01828, G:01832 |
|
1yj9 |
08036 |
Oph·2Pout |
G:0956(OP2) |
|
|
03486, 07591, 07592, 07593, 09187 |
|
A:01006, A:0955 |
|
A:0957, G:0956 |
|
1yj9 |
08068 |
Oph·2Pout |
A:02568(OP2) |
|
|
03566, 04646, 06032, 07670, 09553 |
|
A:02569, G:02567 |
|
A:02569, A:02568 |
|
1yjn |
08020 |
Oph·2Pout |
C:01830(OP1) |
|
|
07750, 07769, 09121, 09124, 09198 |
|
U:01831, A:01842 |
G:01828 |
G:01828, G:01832 |
|
1yjn |
08019 |
Oph·2Pout |
A:02011(OP2) |
|
|
04167, 07233, 07766, 07767, 07768 |
|
A:01941, G:01891 |
U:01890 |
G:01891, C:01892 |
|
1yjn |
08037 |
Oph·2Pout |
A:02553(OP2) |
|
|
03143, 03217, 03661, 07850, 09077 |
|
C:02575, U:02554 |
C:02552 |
A:02576 |
|
1yjw |
78083 |
Oph·2Pout |
A:01754(OP2) |
|
|
07792, 07794, 07797, 07800, 07803 |
|
C:01753, A:01755 |
|
A:01754, A:01755 |
|
1yjw |
78019 |
Oph·2Pout |
A:02011(OP2) |
|
|
08084, 08085, 08123, 08234 |
|
G:01891, A:01941 |
U:01890 |
G:01891 |
|
1yjw |
78020 |
Oph·2Pout |
C:01830(OP1) |
|
|
07913, 07918, 07919, 07921, 07953 |
|
A:01842, U:01831 |
G:01828 |
G:01828, G:01832 |
|
1yjw |
78072 |
Oph·2Pout |
U:0954(OP1) |
|
|
06233, 08673, 08675, 08681, 08684 |
|
C:02298, A:02301 |
|
|
|
1yjw |
78068 |
Oph·2Pout |
A:02568(OP2) |
|
|
09263, 09265, 09268, 09269, E4036 |
|
G:02567, A:02569 |
|
A:02568, A:02569 |
|
1yls |
D341 |
Oph·2Pout |
C:D233(OP2) |
|
|
D394, D395, D396, D397, D398 |
|
G:D208, U:D234 |
|
G:D211, C:D210 |
|
2avy |
A1582 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1791, A1792, A1793, A1794, A1795 |
|
A:A607, A:A609 |
|
C:A631, A:A609, A:A630, A:A608, U:A610 |
|
2avy |
A1593 |
Oph·2Pout |
C:A536(OP1) |
|
|
A1844, A1845, A1846, A1847, A1848 |
|
C:A504, G:A537 |
C:A522 |
|
|
2avy |
A1568 |
Oph·2Pout |
A:A768(OP2) |
|
|
A1727, A1728, A1729, A1730, A1731 |
|
A:A767, U:A804 |
|
G:A769 |
|
2avy |
A1583 |
Oph·2Pout |
A:A1110(OP2) |
|
|
A1796, A1797, A1798, A1799, A1800 |
|
C:A1109, A:A1111 |
|
U:A1189, C:A1112 |
|