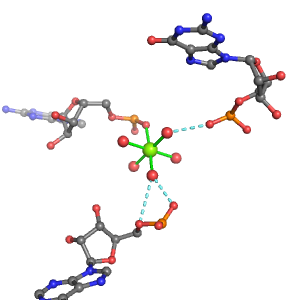
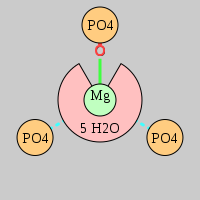
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4tpe |
A1617 |
Oph·2Pout |
A:A560(OP2) |
|
|
A1778, A1779, A1780, A1781, A1782 |
|
U:A562, G:A558 |
A:A559 |
G:A566 |
|
4duz |
A1618 |
Oph·2Pout |
A:A768(OP2) |
|
|
A1937, A1938, A1939, A1940 |
|
C:A805, U:A804 |
|
A:A768, G:A769 |
|
4dr5 |
A1619 |
Oph·2Pout |
A:A768(OP2) |
|
|
A1948, A1949, A1950, A1951 |
|
A:A767, U:A804 |
|
A:A768, G:A769 |
|
4nxn |
A1620 |
Oph·2Pout |
A:A768(OP2) |
|
|
A1942, A1943, A1944, A1945 |
|
C:A805, U:A804 |
|
A:A768, G:A769 |
|
4gd2 |
A1621 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1798, A1799, A1800, A1801, A1802 |
|
A:A607, A:A609 |
|
C:A631, A:A609, A:A630, A:A608, U:A610 |
|
4ton |
A1621 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1795, A1796, A1797, A1798, A1799 |
|
A:A607, A:A609 |
|
A:A609, U:A610, A:A630, A:A608 |
|
4tpe |
A1621 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1797, A1798, A1799, A1800, A1801 |
|
A:A607, A:A609 |
|
C:A631, A:A609, A:A630, A:A608, U:A610 |
|
4tpa |
A1621 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1796, A1797, A1798, A1799, A1800 |
|
A:A607, A:A609 |
|
U:A610, A:A630, C:A631, A:A609 |
|
4pea |
A1621 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1794, A1795, A1796, A1797, A1798 |
|
A:A607, A:A609 |
|
C:A631, A:A609, U:A610, A:A630, A:A608 |
|
4dr2 |
A1621 |
Oph·2Pout |
G:A750(OP2) |
|
|
A2011, A2012, A2013, A2014 |
|
C:A749, A:A653 |
|
G:A752 |
|
4tow |
A1621 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1794, A1795, A1796, A1797, A1798 |
|
A:A607, A:A609 |
|
C:A631, A:A609, U:A610, A:A630, A:A608 |
|
4tp6 |
A1621 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1796, A1797, A1798, A1799, A1800 |
|
A:A607, A:A609 |
|
A:A609, U:A610, A:A630, A:A608 |
|
4gas |
A1621 |
Oph·2Pout |
A:A608(OP2) |
|
|
A1797, A1798, A1799, A1800, A1801 |
|
A:A609, A:A607 |
|
A:A630, C:A631, A:A608, A:A609, U:A610 |
|
4gas |
A1624 |
Oph·2Pout |
A:A814(OP2) |
|
|
A1814, A1815, A1816, A1817, A1818 |
|
U:A904, G:A577 |
|
|
|
4ton |
A1624 |
Oph·2Pout |
A:A814(OP2) |
|
|
A1812, A1813, A1814, A1815, A1816 |
|
A:A815, G:A577 |
|
A:A816 |
|
4tp6 |
A1624 |
Oph·2Pout |
A:A814(OP2) |
|
|
A1813, A1814, A1815, A1816, A1817 |
|
A:A815, G:A577 |
|
A:A816 |
|
4gaq |
A1624 |
Oph·2Pout |
A:A814(OP2) |
|
|
A1812, A1813, A1814, A1815, A1816 |
|
G:A577, A:A815 |
|
A:A816 |
|
4pea |
A1624 |
Oph·2Pout |
A:A814(OP2) |
|
|
A1811, A1812, A1813, A1814, A1815 |
|
A:A815, G:A577 |
|
A:A816 |
|
4tpe |
A1624 |
Oph·2Pout |
A:A814(OP2) |
|
|
A1814, A1815, A1816, A1817, A1818 |
|
A:A815, G:A577 |
|
A:A816 |
|
4tpe |
A1626 |
Oph·2Pout |
U:A891(OP2) |
|
|
A1823, A1824, A1825, A1826, A1827 |
|
A:A892, G:A903 |
|
A:A906, U:A905, U:A904 |
|
4tow |
A1626 |
Oph·2Pout |
U:A891(OP2) |
|
|
A1820, A1821, A1822, A1823, A1824 |
|
A:A892, G:A903 |
|
A:A906, U:A905, U:A904 |
|
4pea |
A1626 |
Oph·2Pout |
U:A891(OP2) |
|
|
A1820, A1821, A1822, A1823, A1824 |
|
A:A892, G:A903 |
|
A:A906, U:A905, U:A904 |
|
4gas |
A1626 |
Oph·2Pout |
U:A891(OP2) |
|
|
A1823, A1824, A1825, A1826, A1827 |
|
A:A892, G:A903 |
|
U:A904, U:A905, G:A903 |
|
4ton |
A1626 |
Oph·2Pout |
U:A891(OP2) |
|
|
A1821, A1822, A1823, A1824, A1825 |
|
A:A892, G:A903 |
|
A:A906, U:A904, U:A905 |
|
4tpa |
A1626 |
Oph·2Pout |
U:A891(OP2) |
|
|
A1822, A1823, A1824, A1825, A1826 |
|
A:A892, G:A903 |
|
A:A906, U:A904, U:A905 |
|