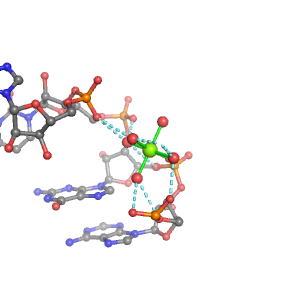
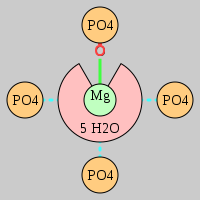
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1m90 |
A8037 |
Oph·3Pout |
A:A2553(OP2) |
|
|
A3177, A7396, A8597, A9657, A9735 |
|
A:A2576, U:A2554, C:A2575 |
C:A2552 |
A:A2576 |
|
1n8r |
A8032 |
Oph·3Pout |
U:A2115(OP1) |
|
|
A3380, A4121, A4184, A4582, C8640 |
|
G:A2272, C:A2273, U:A2116 |
|
|
|
1n8r |
A8045 |
Oph·3Pout |
G:A1979(OP2) |
|
|
A8312, A8423, A8424, A8425 |
|
G:A1998, C:A1999, A:A1978 |
|
|
|
1n8r |
A8017 |
Oph·3Pout |
G:A456(OP2) |
|
|
A7772, A8248, A9809, E8359, E8363 |
|
A:A455, U:A457, G:A458 |
|
|
|
1n8r |
A8035 |
Oph·3Pout |
G:A1484(OP2) |
|
|
A4111, A8293, A8294, A8417, A8418 |
|
C:A1456, A:A1485, C:A1455 |
|
G:A1484, U:A1457 |
|
1n8r |
A8091 |
Oph·3Pout |
G:A918(OP1) |
|
|
A3714, A6870, A8406, A9495, A9937 |
|
C:A2294, C:A2391, A:A2465 |
|
A:A2465 |
|
1n8r |
A8021 |
Oph·3Pout |
G:A2304(OP2) |
|
|
A3353, A4033, A4034, A8254, A8255 |
|
A:A2301, A:A2303, A:A2305 |
|
U:A2306, A:A2305, G:A2304 |
|
1nji |
A8037 |
Oph·3Pout |
A:A2553(OP2) |
|
|
A3132, A3206, A3641, A7858, A9079 |
|
C:A2575, A:A2576, U:A2554 |
C:A2552 |
A:A2576 |
|
1nji |
A8032 |
Oph·3Pout |
U:A2115(OP1) |
|
|
A3716, A3779, A4181, A9984, C8629 |
|
U:A2116, G:A2272, C:A2273 |
|
|
|
1nji |
A8021 |
Oph·3Pout |
G:A2304(OP2) |
|
|
A3631, A3632, A7761, A7762, A9958 |
|
A:A2301, A:A2305, A:A2303 |
|
A:A2305, G:A2304, U:A2306 |
|
1nji |
A8072 |
Oph·3Pout |
U:A954(OP1) |
|
|
A3228, A3987, A7412, A7829, A7888 |
|
C:A2298, G:A2299, A:A2301 |
|
|
|
1q7y |
A8040 |
Oph·3Pout |
U:A392(OP2) |
|
|
A3297, A7340, A7341, A9208 |
|
C:A171, U:A172, U:A391 |
|
|
|
1q7y |
A8058 |
Oph·3Pout |
G:A2013(OP2) |
|
|
A3272, A7296, A7297, C8630, C8632 |
|
G:A1884, U:A1883, C:A2626 |
|
|
|
1q7y |
A8032 |
Oph·3Pout |
U:A2115(OP1) |
|
|
A3235, A3296, A3696, A9506, C8629 |
|
U:A2116, G:A2272, C:A2273 |
|
|
|
1q7y |
A8021 |
Oph·3Pout |
G:A2304(OP2) |
|
|
A3149, A7244, A7245, A9479 |
|
A:A2305, A:A2303, A:A2301 |
|
A:A2305, G:A2304, U:A2306 |
|
1q7y |
A8035 |
Oph·3Pout |
G:A1484(OP2) |
|
|
A3225, A7280, A7281, A7336, A7337 |
|
C:A1455, C:A1456, A:A1485 |
C:A1483 |
G:A1484, U:A1457 |
|
1q81 |
A8037 |
Oph·3Pout |
A:A2553(OP2) |
|
|
A3161, A7347, A8603, A9646, A9725 |
|
A:A2576, U:A2554, C:A2575 |
C:A2552 |
A:A2576 |
|
1q81 |
A8017 |
Oph·3Pout |
G:A456(OP2) |
|
|
A3134, A7246, A8926, E8361, E8464 |
|
A:A455, U:A457, G:A458 |
|
|
|
1q81 |
A8032 |
Oph·3Pout |
U:A2115(OP1) |
|
|
A3238, A3301, A9504, C8565, C8641 |
|
U:A2116, G:A2272, C:A2273 |
G:A2271 |
|
|
1q81 |
A8035 |
Oph·3Pout |
G:A1484(OP2) |
|
|
A3228, A7289, A7290, A7345, A7346 |
|
C:A1456, A:A1485, C:A1455 |
|
G:A1484, U:A1457 |
|
1q81 |
A8040 |
Oph·3Pout |
U:A392(OP2) |
|
|
A4148, A7349, A7350, A9204 |
|
C:A171, U:A172, U:A391 |
|
|
|
1q81 |
A8091 |
Oph·3Pout |
G:A918(OP1) |
|
|
A5984, A7333, A8617, A9060, A9837 |
|
C:A2294, C:A2391, A:A2465 |
|
A:A2465 |
|
1q81 |
A8021 |
Oph·3Pout |
G:A2304(OP2) |
|
|
A3151, A3152, A7252, A7253, A9479 |
|
A:A2301, A:A2305, A:A2303 |
|
A:A2305, G:A2304, U:A2306 |
|
1q82 |
A8058 |
Oph·3Pout |
G:A2013(OP2) |
|
|
A3274, A7296, A7297, C8646, C8648 |
|
G:A1884, U:A1883, C:A2626 |
|
|
|
1q82 |
A8091 |
Oph·3Pout |
G:A918(OP1) |
|
|
A5969, A7323, A8613, A9834 |
|
C:A2294, C:A2391, A:A2465 |
|
A:A2465 |
|