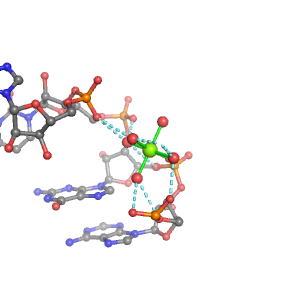
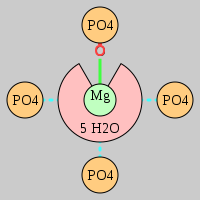
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1s72 |
08017 |
Oph·3Pout |
G:0456(OP2) |
|
|
03465, 07268, 08933, C8356, C8466 |
|
G:0458, A:0455, U:0457 |
|
|
|
1s72 |
08032 |
Oph·3Pout |
U:02115(OP1) |
|
|
03311, 03714, 09512, A8551, A8632 |
|
U:02116, G:02272, C:02273 |
G:02271 |
|
|
1s72 |
08037 |
Oph·3Pout |
A:02553(OP2) |
|
|
03170, 07369, 08598, 09655, 09730 |
|
A:02576, U:02554, C:02575 |
C:02552 |
A:02576 |
|
1vq4 |
08017 |
Oph·3Pout |
G:0456(OP2) |
|
|
03926, 04250, 07932, 09735, C9267 |
|
A:0455, G:0458, U:0457 |
|
|
|
1vq4 |
08021 |
Oph·3Pout |
G:02304(OP2) |
|
|
03278, 03943, 03944, 07938, 07939 |
|
A:02301, A:02303, A:02305 |
|
U:02306, A:02305, G:02304 |
|
1vq4 |
08035 |
Oph·3Pout |
G:01484(OP2) |
|
|
04020, 07975, 07976, 08137, 08138 |
|
C:01455, C:01456, A:01485 |
|
G:01484, U:01457 |
|
1vq4 |
08037 |
Oph·3Pout |
A:02553(OP2) |
|
|
03445, 03517, 03954, 08139, 09397 |
|
C:02575, A:02576, U:02554 |
C:02552 |
A:02576 |
|
1vq4 |
08040 |
Oph·3Pout |
U:0392(OP2) |
|
|
03010, 04094, 04941, 08141, 08142 |
|
U:0172, U:0391, C:0171 |
|
|
|
1vq4 |
08032 |
Oph·3Pout |
U:02115(OP1) |
|
|
03304, 04093, 04499, A9347, A9421 |
|
C:02273, U:02116, G:02272 |
|
|
|
1vq4 |
08036 |
Oph·3Pout |
G:0956(OP2) |
|
|
03975, 07977, 07978, 07979, 09688 |
|
A:01006, A:0955, A:0957 |
|
A:0957, G:0956 |
|
1vq5 |
08040 |
Oph·3Pout |
U:0392(OP2) |
|
|
03017, 04104, 04949, 08186, 08187 |
|
U:0172, U:0391, C:0171 |
|
|
|
1vq5 |
08032 |
Oph·3Pout |
U:02115(OP1) |
|
|
03311, 04103, 04504, A9343, A9422 |
|
C:02273, U:02116, G:02272 |
|
|
|
1vq5 |
08036 |
Oph·3Pout |
G:0956(OP2) |
|
|
03986, 08128, 08129, 08130, 09693 |
|
A:01006, A:0955, A:0957 |
|
A:0957, G:0956 |
|
1vq5 |
08021 |
Oph·3Pout |
G:02304(OP2) |
|
|
03286, 03954, 03955, 07983, 07984 |
|
A:02301, A:02303, A:02305 |
|
U:02306, A:02305, G:02304 |
|
1vq5 |
08017 |
Oph·3Pout |
G:0456(OP2) |
|
|
04257, 07977, 09738, C9155, C9261 |
|
G:0458, A:0455, U:0457 |
|
|
|
1vq5 |
08037 |
Oph·3Pout |
A:02553(OP2) |
|
|
03457, 03532, 03965, 08184, 09399 |
|
C:02575, A:02576, U:02554 |
C:02552 |
A:02576 |
|
1vq6 |
08032 |
Oph·3Pout |
U:02115(OP1) |
|
|
03300, 04095, 04496, A9345, A9425 |
|
C:02273, U:02116, G:02272 |
|
|
|
1vq6 |
08017 |
Oph·3Pout |
G:0456(OP2) |
|
|
03929, 04249, 07941, 09736, C9263 |
|
A:0455, G:0458, U:0457 |
|
|
|
1vq6 |
08036 |
Oph·3Pout |
G:0956(OP2) |
|
|
03978, 07985, 07986, 07987, 09690 |
|
A:01006, A:0955, A:0957 |
|
A:0957, G:0956 |
|
1vq6 |
08021 |
Oph·3Pout |
G:02304(OP2) |
|
|
03275, 03946, 03947, 07947, 07948 |
|
A:02301, A:02305, A:02303 |
|
U:02306, A:02305, G:02304 |
|
1vq7 |
08021 |
Oph·3Pout |
G:02304(OP2) |
|
|
03286, 03962, 03963, 07978, 07979 |
|
A:02301, A:02305, A:02303 |
|
U:02306, A:02305, G:02304 |
|
1vq7 |
08036 |
Oph·3Pout |
G:0956(OP2) |
|
|
03994, 08124, 08125, 08126, 09693 |
|
A:01006, A:0955, A:0957 |
|
A:0957, G:0956 |
|
1vq7 |
08017 |
Oph·3Pout |
G:0456(OP2) |
|
|
04268, 07622, 07972, 09740, C9155 |
|
G:0458, A:0455, U:0457 |
|
|
|
1vq7 |
08032 |
Oph·3Pout |
U:02115(OP1) |
|
|
03312, 04112, 04516, A9349, A9424 |
|
C:02273, U:02116, G:02272 |
|
|
|
1vq7 |
08037 |
Oph·3Pout |
A:02553(OP2) |
|
|
03459, 03533, 03973, 08179, 09398 |
|
C:02575, A:02576, U:02554 |
C:02552 |
A:02576 |
|