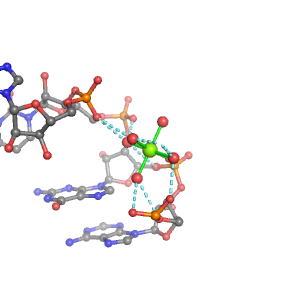
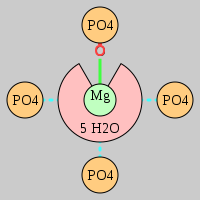
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4qct |
A3287 |
Oph·3Pout |
A:A2062(OP2) |
|
|
A4122, A4123, A4124, A4125, A4126 |
|
G:A2444, C:A2443, G:A2061 |
|
G:A2444, G:A2445 |
|
4qcs |
A3096 |
Oph·3Pout |
C:A578(OP1) |
|
|
A4056, A4057, A4058, A4059, A4060 |
|
U:A820, C:A817, G:A576 |
|
|
|
4qcr |
A3565 |
Oph·3Pout |
G:A1859(OP2) |
|
|
A4842, A4843, A4844, A4845, A4846 |
|
C:A1858, A:A1994, C:A1819 |
|
G:A1859 |
|
4qcr |
A3427 |
Oph·3Pout |
U:A2096(OP1) |
|
|
A4427, A4428, A4429, D401 |
|
G:A2251, C:A2252, U:A2097 |
G:A2250 |
|
|
4qcr |
A3540 |
Oph·3Pout |
G:A586(OP2) |
|
|
A4751, A4752, A4753, A4754, A4755 |
|
C:A587, A:A595, G:A1021 |
C:A1020 |
|
|
4qcr |
A3347 |
Oph·3Pout |
U:A2039(OP1) |
|
|
A4190, A4191, A4192, A4193, A4194 |
|
A:A601, G:A602, G:A2040 |
|
|
|
4qcr |
A3408 |
Oph·3Pout |
U:A2418(OP1) |
|
|
A4348, A4349, A4350, A4351, A4352 |
|
G:A2419, U:A2420, G:A2421 |
G:A2417 |
G:A2422, U:A2420, G:A2421 |
|
4qcr |
A3307 |
Oph·3Pout |
U:A1008(OP1) |
|
|
A4073, A4074, A4075, A4076, A4077 |
|
C:A2510, A:A2509, G:A992 |
|
|
|
4qcr |
A3512 |
Oph·3Pout |
C:A2464(OP1) |
|
|
A4664, A4665, A4666, A4667, A5380 |
|
U:A2505, G:A2506, A:A2465 |
|
|
|
4qcr |
A3515 |
Oph·3Pout |
C:A1463(OP2) |
|
|
A4672, A4673, A4674, A4675 |
|
G:A1464, C:A1624, U:A1625 |
|
G:A1464 |
|
4qcr |
A3348 |
Oph·3Pout |
A:A2084(OP2) |
|
|
A4195, A4196, A4197, A4198, A4199 |
|
G:A2456, C:A2455, G:A2083 |
|
G:A2456, G:A2457 |
|
4qcr |
A3410 |
Oph·3Pout |
C:A238(OP1) |
|
|
A4357, A4358, A4359, A4360, A4361 |
|
G:A239, U:A2443, A:A2444 |
G:A239 |
C:A2406 |
|
4qcq |
A3125 |
Oph·3Pout |
A:A768(OP2) |
|
|
A4041, A4042, A4043, A4044, A4045 |
|
U:A804, C:A805, A:A767 |
|
A:A768, G:A769 |
|
4qcp |
A3429 |
Oph·3Pout |
G:A1647(OP2) |
|
|
A4493, A4494, A4495, A4496, A4497 |
|
G:A1325, A:A1272, C:A1617 |
A:A1616, C:A1646 |
A:A1616 |
|
4qcp |
A3395 |
Oph·3Pout |
C:A1417(OP2) |
|
|
A4402, A4403, A4404, A4405 |
|
C:A1577, U:A1578, G:A1418 |
G:A1416 |
G:A1418 |
|
4qcp |
A3441 |
Oph·3Pout |
G:A1828(OP2) |
|
|
A4526, A4527, A4528, A4529, D413 |
|
C:A1827, A:A1972, C:A1788 |
|
G:A1828 |
|
4qcp |
A3251 |
Oph·3Pout |
U:A963(OP1) |
|
|
A4040, A4041, A4042, A4043, A4044 |
|
C:A2498, A:A2497, G:A947 |
|
|
|
4qcp |
A3282 |
Oph·3Pout |
A:A2062(OP2) |
|
|
A4121, A4122, A4123, A4124, A4125 |
|
G:A2444, C:A2443, G:A2061 |
|
G:A2444, G:A2445 |
|
4qcp |
A3578 |
Oph·3Pout |
C:A2452(OP1) |
|
|
A4835, A4836, A4837, A4838, 0201 |
|
A:A2453, G:A2494, U:A2493 |
|
|
|
4qcp |
A3579 |
Oph·3Pout |
C:A1636(OP2) |
|
|
A4839, A4840, A4841, A4842, A4843 |
|
U:A1300, G:A1635, A:A1637 |
U:A1300 |
|
|
4qcn |
A3542 |
Oph·3Pout |
G:A1859(OP2) |
|
|
A4820, A4821, A4822, A4823, A4824 |
|
C:A1858, A:A1994, C:A1819 |
|
G:A1859 |
|
4qcn |
A3404 |
Oph·3Pout |
U:A2096(OP1) |
|
|
A4405, A4406, A4407, D3102 |
|
C:A2252, U:A2097, G:A2251 |
G:A2250 |
|
|
4qcn |
A3504 |
Oph·3Pout |
U:A2504(OP1) |
|
|
A4696, A4697, A4698, A4699, A4700 |
|
U:A2505, A:A2465, G:A2466 |
C:A2585 |
C:A2585 |
|
4qcn |
A3488 |
Oph·3Pout |
C:A2464(OP1) |
|
|
A4645, A4646, A4647, 03102, 03103 |
|
U:A2505, G:A2506, A:A2465 |
|
|
|
4qcn |
A3328 |
Oph·3Pout |
U:A2039(OP1) |
|
|
A4177, A4178, A4179, A4180, A4181 |
|
A:A601, G:A602, G:A2040 |
|
|
|