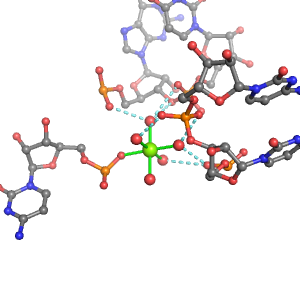
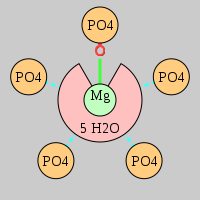
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3la5 |
A11 |
Oph·4Pout |
G:A44(OP1) |
|
|
A9, A191, A302, A411, A412 |
|
C:A50, G:A44, C:A51, A:A45 |
|
|
|
3or9 |
A1540 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1608, A1609, A1610, A1611, A1612 |
|
U:A562, A:A563, G:A557, G:A558 |
|
G:A566 |
|
3i1m |
A1540 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1608, A1609, A1610, A1611, A1612 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
3oaq |
A1540 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1608, A1609, A1610, A1611, A1612 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
3ofx |
A1540 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1607, A1608, A1609, A1610, A1611 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
2qbj |
A1547 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1623, A1624, A1625, A1626, A1627 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
|
|
2z4m |
A1548 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1623, A1624, A1625, A1626, A1627 |
|
U:A562, A:A563, G:A558, G:A557 |
A:A559 |
|
|
2qbh |
A1549 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1635, A1636, A1637, A1638, A1639 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
2qou |
A1549 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1928, A1929, A1930, A1931, A1932 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
|
|
2qoy |
A1549 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1930, A1931, A1932, A1933, A1934 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
|
|
2z4k |
A1550 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1635, A1636, A1637, A1638, A1639 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
3i1s |
A1559 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1713, A1714, A1715, A1716 |
|
A:A964, U:A965, U:A1199, C:A1054 |
|
|
|
3i1o |
A1559 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1713, A1714, A1715, A1716 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3i21 |
A1559 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1713, A1714, A1715, A1716 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3oar |
A1561 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1711, A1712, A1713, A1714 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3ofb |
A1562 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1711, A1712, A1713, A1714 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3ofy |
A1562 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1711, A1712, A1713, A1714 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3ora |
A1562 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1717, A1718, A1719, A1720 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3ofp |
A1562 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1709, A1710, A1711, A1712 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
2qp0 |
A1576 |
Oph·4Pout |
C:A536(OP1) |
|
|
A2047, A2048, A2049, A2050, A2051 |
|
C:A504, G:A537, A:A523, C:A503 |
C:A522 |
|
|
4tou |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
4tp4 |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A557, G:A558 |
|
G:A566 |
|
4tp0 |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A557, G:A558 |
|
G:A566 |
|
4tol |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A558, G:A557 |
|
|
|
4tpc |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A557, G:A558 |
|
G:A566 |
|