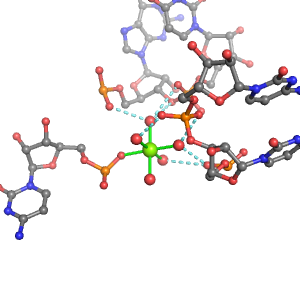
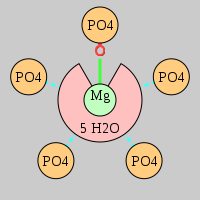
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1vq7 |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
04039, 08122, 08123, 08177, 08178 |
|
C:01456, C:01483, A:01485, C:01455 |
|
G:01484, U:01457 |
|
1vq8 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03256, 03828, 04240, 08380, 09665 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
1vqk |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03264, 03837, 04251, 08418, 09665 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
1vql |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03256, 03827, 04239, 08384, 09664 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
1vqm |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03258, 03833, 04246, 08385, 09664 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
1vqn |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03258, 03830, 04243, 08377, 09665 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
1vqo |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03269, 03849, 04264, 08433, 09664 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
1vqp |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03265, 03841, 04259, 08405, 09664 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
1vs6 |
B2917 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3072, B3073, B3074, B3075, B3076 |
|
C:B1314, G:B1332, G:B1334, U:B1313 |
|
G:B1334, G:B1333 |
|
1vs6 |
B2992 |
Oph·4Pout |
C:B31(OP1) |
|
|
B3421, B3422, B3423, B3424, B3425 |
|
G:B1239, U:B1240, C:B1200, U:B1199 |
|
|
|
1vt2 |
A3019 |
Oph·4Pout |
U:A2243(OP1) |
|
|
A3564, A3565, A3566, A3567 |
|
C:A192, A:A199, C:A198, U:A193 |
|
|
|
1vt2 |
A2961 |
Oph·4Pout |
G:A1828(OP1) |
|
|
A3306, A3307, A3308, A3309, A3310 |
|
A:A1773, G:A1972, G:A1973, A:A1772 |
|
|
|
1vt2 |
A3010 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3531, A3532, A3533, A3534, A3535 |
|
G:A1239, C:A1200, U:A1199, U:A1240 |
|
|
|
1yhq |
08008 |
Oph·4Pout |
G:028(OP1) |
|
|
03139, 03729, 04144, 08368, 09537 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
1yhq |
08070 |
Oph·4Pout |
G:0918(OP1) |
|
|
03049, 03824, 06950, 08362, 09588 |
|
A:02465, G:02293, C:02294, C:02391 |
|
A:02465 |
|
1yi2 |
08091 |
Oph·4Pout |
G:0918(OP1) |
|
|
03325, 06465, 07809, 09091, 09549 |
|
G:02293, C:02294, A:02465, U:0919 |
|
A:02465 |
|
1yi2 |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
03713, 07767, 07768, 07822, 07823 |
|
C:01456, C:01483, A:01485, C:01455 |
|
G:01484, U:01457 |
|
1yi2 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03229, 03645, 07815, 09037, 09639 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
1yij |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03236, 03657, 07828, 09038, 09640 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
1yj9 |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
03530, 07589, 07590, 07644 |
|
C:01456, C:01483, A:01485, C:01455 |
|
G:01484, U:01457 |
|
1yjn |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
03729, 07792, 07793, 07848, 07849 |
|
C:01483, C:01455, C:01456, A:01485 |
|
U:01457 |
|
1yjw |
78091 |
Oph·4Pout |
G:0918(OP1) |
|
|
06150, 08660, 08668, 09021, 09022 |
|
C:02294, A:02465, C:02391, U:0919 |
|
G:02397, A:02465 |
|
2aw4 |
B2917 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3069, B3070, B3071, B3072, B3073 |
|
C:B1314, G:B1332, G:B1334, U:B1313 |
|
G:B1334, G:B1333 |
|
2aw4 |
B2992 |
Oph·4Pout |
C:B31(OP1) |
|
|
B3417, B3418, B3419, B3420, B3421 |
|
G:B1239, U:B1240, C:B1200, U:B1199 |
|
|
|
2awb |
B3014 |
Oph·4Pout |
U:B963(OP1) |
|
|
B3509, B3510, B3511, B3512, B3513 |
|
A:B947, A:B2497, C:B964, C:B2498 |
|
|
|