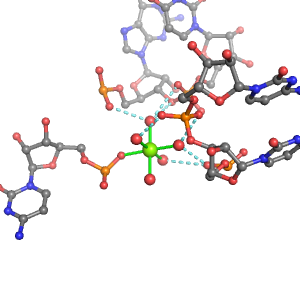
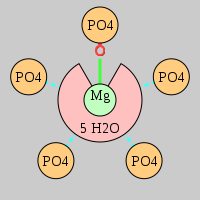
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2i2t |
B2920 |
Oph·4Pout |
G:B1828(OP1) |
|
|
B3094, B3095, B3096, B3097, B3098 |
|
A:B1773, G:B1972, G:B1973, A:B1772 |
|
|
|
2i2t |
B2917 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3079, B3080, B3081, B3082, B3083 |
|
C:B1314, G:B1332, G:B1334, U:B1313 |
|
G:B1334, G:B1333, C:B1335 |
|
2i2t |
B2992 |
Oph·4Pout |
C:B31(OP1) |
|
|
B3427, B3428, B3429, B3430, B3431 |
|
G:B1239, U:B1240, U:B1199, C:B1200 |
|
|
|
2i2v |
B2979 |
Oph·4Pout |
G:B1828(OP1) |
|
|
B3383, B3384, B3385, B3386, B3387 |
|
G:B1972, G:B1973, A:B1773, A:B1772 |
|
|
|
2i2v |
B2975 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3366, B3367, B3368, B3369, B3370 |
|
C:B1314, G:B1334, U:B1313, G:B1332 |
|
G:B1334, C:B1335 |
|
2otj |
08091 |
Oph·4Pout |
G:0918(OP1) |
|
|
03338, 06506, 07845, 09092, 09558 |
|
C:02294, C:02391, A:02465, U:0919 |
|
A:02465 |
|
2otl |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03236, 03659, 07838, 09040, 09637 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
2qal |
A2038 |
Oph·4Pout |
A:A560(OP2) |
|
|
A2039, A2040, A2041, A2042, A2043 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
2qam |
B3499 |
Oph·4Pout |
C:B31(OP1) |
|
|
B4018, B4019, B4020, B4021, B4022 |
|
G:B1239, U:B1199, C:B1200, U:B1240 |
|
|
|
2qan |
A2023 |
Oph·4Pout |
A:A560(OP2) |
|
|
A2024, A2025, A2026, A2027, A2028 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
2qba |
B3499 |
Oph·4Pout |
C:B31(OP1) |
|
|
B4015, B4016, B4017, B4018, B4019 |
|
G:B1239, U:B1240, C:B1200, U:B1199 |
|
|
|
2qbb |
A2023 |
Oph·4Pout |
A:A560(OP2) |
|
|
A2388, A2389, A2390, A2391, A2392 |
|
U:A562, A:A563, G:A558, G:A557 |
A:A559 |
G:A566 |
|
2qbc |
B3515 |
Oph·4Pout |
C:B31(OP1) |
|
|
B4037, B4038, B4039, B4040, B4041 |
|
C:B1200, G:B1239, U:B1240, U:B1199 |
|
|
|
2qbc |
B3425 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3967, B3968, B3969, B3970, B3971 |
|
C:B1314, G:B1332, G:B1334, U:B1313 |
|
G:B1334, G:B1333 |
|
2qbd |
A2038 |
Oph·4Pout |
A:A560(OP2) |
|
|
A2039, A2040, A2041, A2042, A2043 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
2qbe |
B3072 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3669, B3670, B3671, B3672, B3673 |
|
C:B1314, G:B1332, G:B1334, U:B1313 |
|
G:B1333, C:B1335 |
|
2qbe |
B3499 |
Oph·4Pout |
C:B31(OP1) |
|
|
B4015, B4016, B4017, B4018, B4019 |
|
G:B1239, C:B1200, U:B1199, U:B1240 |
A:B1214 |
|
|
2qbh |
A1549 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1635, A1636, A1637, A1638, A1639 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
2qbi |
B2920 |
Oph·4Pout |
G:B1828(OP1) |
|
|
B3085, B3086, B3087, B3088, B3089 |
|
A:B1773, G:B1972, G:B1973, A:B1772 |
|
|
|
2qbi |
B2992 |
Oph·4Pout |
C:B31(OP1) |
|
|
B3417, B3418, B3419, B3420, B3421 |
|
G:B1239, U:B1240, C:B1200, U:B1199 |
|
|
|
2qbj |
A1547 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1623, A1624, A1625, A1626, A1627 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
|
|
2qbk |
B2976 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3363, B3364, B3365, B3366, B3367 |
|
C:B1314, G:B1332, G:B1334, U:B1313 |
|
G:B1333 |
|
2qbk |
B3015 |
Oph·4Pout |
U:B963(OP1) |
|
|
B3511, B3512, B3513, B3514, B3515 |
|
C:B964, C:B2498, A:B2497, A:B947 |
|
|
|
2qex |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03059, 03472, 07690, 08862, 09461 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
2qou |
A1549 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1928, A1929, A1930, A1931, A1932 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
|
|