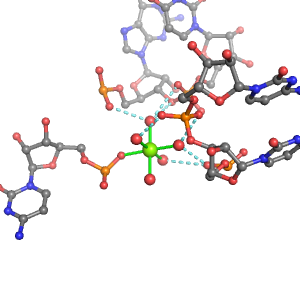
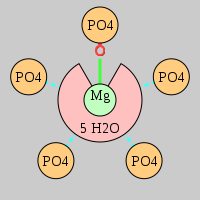
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2qox |
B2975 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3361, B3362, B3363, B3364, B3365 |
|
C:B1314, G:B1334, U:B1313, G:B1332 |
|
G:B1334, G:B1333 |
|
2qoy |
A1549 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1930, A1931, A1932, A1933, A1934 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
|
|
2qoz |
B2920 |
Oph·4Pout |
G:B1828(OP1) |
|
|
B3580, B3581, B3582, B3583, B3584 |
|
A:B1773, G:B1972, G:B1973, A:B1772 |
|
|
|
2qp0 |
A1576 |
Oph·4Pout |
C:A536(OP1) |
|
|
A2047, A2048, A2049, A2050, A2051 |
|
C:A504, G:A537, A:A523, C:A503 |
C:A522 |
|
|
2qp1 |
B2976 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B3870, B3871, B3872, B3873, B3874 |
|
C:B1314, G:B1332, G:B1334, U:B1313 |
|
G:B1334, G:B1333 |
|
2qp1 |
B2994 |
Oph·4Pout |
C:B31(OP1) |
|
|
B3940, B3941, B3942, B3943, B3944 |
|
C:B1200, U:B1199, G:B1239, U:B1240 |
|
|
|
2z4k |
A1550 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1635, A1636, A1637, A1638, A1639 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
2z4l |
B2993 |
Oph·4Pout |
C:B31(OP1) |
|
|
B3419, B3420, B3421, B3422, B3423 |
|
G:B1239, C:B1200, U:B1199, U:B1240 |
|
|
|
2z4m |
A1548 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1623, A1624, A1625, A1626, A1627 |
|
U:A562, A:A563, G:A558, G:A557 |
A:A559 |
|
|
2z4n |
B3016 |
Oph·4Pout |
U:B963(OP1) |
|
|
B3515, B3516, B3517, B3518, B3519 |
|
A:B947, C:B964, C:B2498, A:B2497 |
|
|
|
3cc2 |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
03269, 07369, 07370, 07425, 07426 |
|
C:01456, C:01483, A:01485, C:01455 |
|
G:01484, U:01457 |
|
3cc2 |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03201, 07418, 08560, 09173, 09772 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
3ccm |
08008 |
Oph·4Pout |
G:028(OP1) |
|
|
03251, 03676, 07889, 09047, 09660 |
|
C:01305, U:01304, G:01344, C:01343 |
|
|
|
3ccq |
08008 |
Oph·4Pout |
G:028(OP1) |
|
|
03250, 03669, 07906, 09049, 09653 |
|
G:01344, C:01305, U:01304, C:01343 |
|
|
|
3ccr |
08008 |
Oph·4Pout |
G:028(OP1) |
|
|
03131, 03657, 07849, 09047, 09654 |
|
C:01305, U:01304, G:01344, C:01343 |
|
|
|
3ccs |
08008 |
Oph·4Pout |
G:028(OP1) |
|
|
03228, 03648, 07858, 09046, 09646 |
|
C:01305, U:01304, C:01343, G:01344 |
A:01318 |
|
|
3ccv |
08008 |
Oph·4Pout |
G:028(OP1) |
|
|
03236, 03657, 07864, 09044, 09645 |
|
C:01305, U:01304, C:01343, G:01344 |
|
|
|
3cpw |
08008 |
Oph·4Pout |
G:028(OP1) |
|
|
03101, 03678, 04085, 08282, 09535 |
|
C:01343, G:01344, C:01305, U:01304 |
|
|
|
3cxc |
08035 |
Oph·4Pout |
G:01484(OP2) |
|
|
04664, 08702, 08703, 08756, 08757 |
|
C:01456, C:01483, A:01485, C:01455 |
|
G:01484, U:01457 |
|
3cxc |
08012 |
Oph·4Pout |
G:028(OP1) |
|
|
03037, 03617, 04190, 04596, 08750 |
|
U:01304, C:01305, C:01343, G:01344 |
|
|
|
3df1 |
A2038 |
Oph·4Pout |
A:A560(OP2) |
|
|
A4039, A4040, A4041, A4042, A4043 |
|
A:A563, U:A562, G:A557, G:A558 |
|
G:A566 |
|
3df2 |
B3072 |
Oph·4Pout |
G:B1333(OP2) |
|
|
B4073, B4074, B4075, B4076, B4077 |
|
C:B1314, G:B1334, U:B1313, G:B1332 |
|
G:B1334, C:B1335 |
|
3df2 |
B3499 |
Oph·4Pout |
C:B31(OP1) |
|
|
B4500, B4501, B4502, B4503, B4504 |
|
G:B1239, U:B1240, C:B1200, U:B1199 |
|
|
|
3df3 |
A2023 |
Oph·4Pout |
A:A560(OP2) |
|
|
A4024, A4025, A4026, A4027, A4028 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
3i1m |
A1540 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1608, A1609, A1610, A1611, A1612 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|