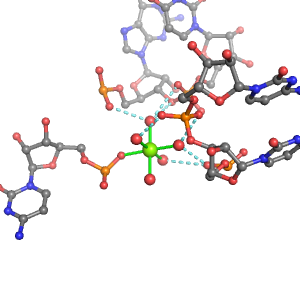
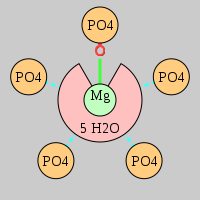
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3ofd |
A2960 |
Oph·4Pout |
G:A1828(OP1) |
|
|
A3303, A3304, A3305, A3306, C338 |
|
A:A1772, A:A1773, G:A1972, G:A1973 |
|
|
|
3ofd |
A3018 |
Oph·4Pout |
U:A2243(OP1) |
|
|
A3560, A3561, A3562, A3563 |
|
C:A192, A:A199, C:A198, U:A193 |
|
|
|
3ofd |
A3009 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3527, A3528, A3529, A3530, A3531 |
|
G:A1239, C:A1200, U:A1199, U:A1240 |
A:A1214 |
|
|
3ofp |
A1562 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1709, A1710, A1711, A1712 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3ofq |
A2948 |
Oph·4Pout |
G:A1333(OP2) |
|
|
A3244, A3245, A3246, A3247, A3248 |
|
C:A1314, G:A1332, G:A1334, U:A1313 |
G:A1331 |
G:A1333 |
|
3ofq |
A3019 |
Oph·4Pout |
U:A2243(OP1) |
|
|
A3560, A3561, A3562, A3563 |
|
C:A192, A:A199, C:A198, U:A193 |
|
|
|
3ofq |
A3010 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3526, A3527, A3528, A3529, A3530 |
|
G:A1239, C:A1200, U:A1199, U:A1240 |
A:A1214 |
|
|
3ofr |
A3013 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3539, A3540, A3541, A3542, A3543 |
|
C:A1200, U:A1240, G:A1239, U:A1199 |
|
|
|
3ofr |
A2999 |
Oph·4Pout |
U:A1636(OP2) |
|
|
A3485, A3486, A3487, A3488, A3489 |
|
A:A1301, A:A1637, G:A1300, G:A1299 |
|
|
|
3ofx |
A1540 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1607, A1608, A1609, A1610, A1611 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
3ofy |
A1562 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1711, A1712, A1713, A1714 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3og0 |
A3017 |
Oph·4Pout |
U:A2243(OP1) |
|
|
A3559, A3560, A3561, A3562 |
|
C:A192, A:A199, C:A198, U:A193 |
|
|
|
3og0 |
A2959 |
Oph·4Pout |
G:A1828(OP1) |
|
|
A3303, A3304, A3305, A3306, C338 |
|
A:A1772, A:A1773, G:A1972, G:A1973 |
|
|
|
3og0 |
A2946 |
Oph·4Pout |
G:A1333(OP2) |
|
|
A3241, A3242, A3243, A3244, A3245 |
|
C:A1314, G:A1332, G:A1334, U:A1313 |
G:A1331 |
C:A1335 |
|
3og0 |
A2937 |
Oph·4Pout |
U:A963(OP1) |
|
|
A3203, A3204, A3205, A3206, A3207 |
|
C:A964, C:A2498, A:A2497, A:A947 |
|
|
|
3og0 |
A3008 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3527, A3528, A3529, A3530, A3531 |
|
G:A1239, C:A1200, U:A1199, U:A1240 |
A:A1214 |
|
|
3or9 |
A1540 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1608, A1609, A1610, A1611, A1612 |
|
U:A562, A:A563, G:A557, G:A558 |
|
G:A566 |
|
3ora |
A1562 |
Oph·4Pout |
G:A1198(OP1) |
|
|
A1717, A1718, A1719, A1720 |
|
A:A964, U:A965, C:A1054, U:A1199 |
|
|
|
3orb |
A3012 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3543, A3544, A3545, A3546, A3547 |
|
C:A1200, G:A1239, U:A1240, U:A1199 |
|
|
|
3r8s |
A2946 |
Oph·4Pout |
G:A1333(OP2) |
|
|
A3291, A3292, A3293, A3294, A3295 |
|
C:A1314, G:A1334, U:A1313, G:A1332 |
|
G:A1334, G:A1333, C:A1335 |
|
3r8s |
A3014 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3607, A3608, A3609, A3610, A3611 |
|
U:A1240, C:A1200, G:A1239, U:A1199 |
|
|
|
3r8s |
A2959 |
Oph·4Pout |
G:A1828(OP1) |
|
|
A3353, A3354, A3355, A3356, A3357 |
|
A:A1773, G:A1972, G:A1973, A:A1772 |
|
|
|
3r8t |
A2945 |
Oph·4Pout |
G:A1333(OP2) |
|
|
A3264, A3265, A3266, A3267, A3268 |
|
C:A1314, G:A1332, G:A1334, U:A1313 |
|
G:A1334, G:A1333, C:A1335 |
|
3v22 |
A1705 |
Oph·4Pout |
U:A560(OP2) |
|
|
A1935, A1936, A1937, A1938 |
|
C:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
3v25 |
A3267 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3617, A3618, A3619, A3620 |
|
U:A1199, C:A1200, U:A1240, G:A1239 |
|
|
|