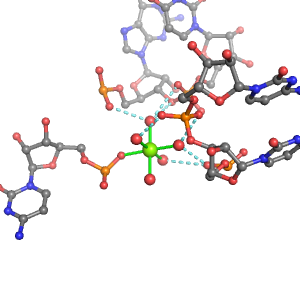
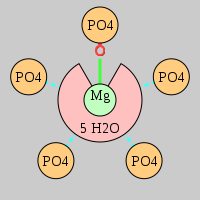
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4qct |
A3595 |
Oph·4Pout |
C:A1636(OP2) |
|
|
A4861, A4862, A4863, A4864, A4865 |
|
G:A1299, U:A1300, G:A1635, A:A1637 |
U:A1300 |
|
|
4qcv |
A3517 |
Oph·4Pout |
G:A1859(OP1) |
|
|
A4638, A4639, A4640, A4641, A4642 |
|
G:A1803, A:A1994, A:A1804, G:A1995 |
|
|
|
4qcv |
A3535 |
Oph·4Pout |
C:A31(OP1) |
|
|
A4686, A4687, A4688, A4689 |
|
G:A1285, U:A1244, C:A1245, U:A1286 |
|
|
|
4qcx |
A3337 |
Oph·4Pout |
G:A741(OP1) |
|
|
A4188, A4189, A4190, A4191 |
|
A:A1784, A:A1785, C:A1983, G:A2608 |
G:A2607 |
|
|
4qcz |
A3546 |
Oph·4Pout |
G:A1859(OP1) |
|
|
A4757, A4758, A4759, A4760, A4761 |
|
G:A1803, A:A1994, A:A1804, G:A1995 |
|
|
|
4qd1 |
A3454 |
Oph·4Pout |
G:A1828(OP1) |
|
|
A4420, A4421, A4422, A4423 |
|
G:A1772, A:A1972, A:A1773, G:A1973 |
|
|
|
4qd1 |
A3584 |
Oph·4Pout |
C:A1636(OP2) |
|
|
A4687, A4688, A4689, A4690, A4691 |
|
G:A1299, U:A1300, G:A1635, A:A1637 |
|
|
|
4qd1 |
A3282 |
Oph·4Pout |
U:A963(OP1) |
|
|
A4026, A4027, A4028, A4029, A4030 |
|
C:A964, C:A2498, A:A2497, G:A947 |
|
|
|
4tol |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A558, G:A557 |
|
|
|
4ton |
A1617 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1776, A1777, A1778, A1779, A1780 |
|
U:A562, A:A563, G:A558, G:A557 |
A:A559 |
G:A566 |
|
4too |
A3121 |
Oph·4Pout |
U:A2243(OP1) |
|
|
A3733, A3734, A3735, A3736 |
|
C:A192, U:A193, C:A198, A:A199 |
|
|
|
4tou |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
4tov |
A3098 |
Oph·4Pout |
U:A1636(OP2) |
|
|
A3650, A3651, A3652, A3653, A3654 |
|
G:A1299, A:A1637, A:A1635, G:A1300 |
|
|
|
4tox |
A3044 |
Oph·4Pout |
G:A1333(OP2) |
|
|
A3388, A3389, A3390, A3391, A3392 |
|
U:A1313, C:A1314, G:A1334, G:A1332 |
|
G:A1334, C:A1335 |
|
4tox |
A3112 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3699, A3700, A3701, A3702, A3703 |
|
U:A1199, G:A1239, U:A1240, C:A1200 |
|
|
|
4tox |
A3121 |
Oph·4Pout |
U:A2243(OP1) |
|
|
A3738, A3739, A3740, A3741 |
|
C:A192, U:A193, C:A198, A:A199 |
|
|
|
4tp0 |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A557, G:A558 |
|
G:A566 |
|
4tp1 |
A3112 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3703, A3704, A3705, A3706, A3707 |
|
C:A1200, G:A1239, U:A1240, U:A1199 |
A:A1214 |
|
|
4tp2 |
A1617 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1777, A1778, A1779, A1780, A1781 |
|
U:A562, A:A563, G:A557, G:A558 |
A:A559 |
G:A566 |
|
4tp3 |
A3043 |
Oph·4Pout |
G:A1333(OP2) |
|
|
A3388, A3389, A3390, A3391, A3392 |
|
C:A1314, G:A1332, G:A1334, U:A1313 |
|
G:A1333, C:A1335 |
|
4tp3 |
A3097 |
Oph·4Pout |
U:A1636(OP2) |
|
|
A3642, A3643, A3644, A3645, A3646 |
|
A:A1301, A:A1635, A:A1637, G:A1300 |
|
|
|
4tp4 |
A1607 |
Oph·4Pout |
A:A560(OP2) |
|
|
A1731, A1732, A1733, A1734, A1735 |
|
U:A562, A:A563, G:A557, G:A558 |
|
G:A566 |
|
4tp5 |
A3111 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3705, A3706, A3707, A3708, A3709 |
|
C:A1200, G:A1239, U:A1199, U:A1240 |
A:A1214 |
|
|
4tp7 |
A3057 |
Oph·4Pout |
G:A1828(OP1) |
|
|
A3447, A3448, A3449, A3450, A3451 |
|
A:A1773, G:A1972, G:A1973, A:A1772 |
|
|
|
4tp9 |
A3112 |
Oph·4Pout |
C:A31(OP1) |
|
|
A3701, A3702, A3703, A3704, A3705 |
|
C:A1200, G:A1239, U:A1240, U:A1199 |
A:A1214 |
|
|