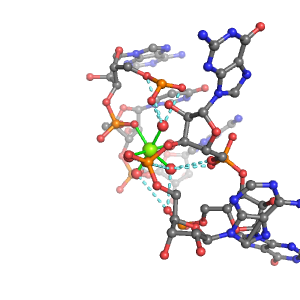
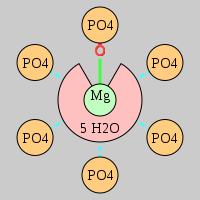
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3ofz |
A2997 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3477, A3478, A3479, A3480, A3481 |
|
A:A1637, A:A1301, A:A1635, G:A1299, G:A1300 |
|
|
|
3ofc |
A2998 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3488, A3489, A3490, A3491, A3492 |
|
G:A1300, A:A1637, A:A1301, A:A1635, G:A1299 |
|
|
|
3r8t |
A2999 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3521, A3522, A3523, A3524, A3525 |
|
A:A1301, A:A1637, A:A1635, G:A1299, G:A1300 |
|
|
|
3i1r |
A3000 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3494, A3495, A3496, A3497, A3498 |
|
A:A1301, G:A1300, A:A1637, A:A1635, G:A1299 |
|
|
|
3r8s |
A3000 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3550, A3551, A3552, A3553, A3554 |
|
A:A1301, A:A1637, A:A1635, G:A1299, G:A1300 |
|
|
|
3i20 |
A3000 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3494, A3495, A3496, A3497, A3498 |
|
A:A1301, A:A1637, A:A1635, G:A1299, G:A1300 |
|
|
|
4pec |
A3096 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3643, A3644, A3645, A3646, A3647 |
|
A:A1301, A:A1637, G:A1299, A:A1635, G:A1300 |
|
|
|
4too |
A3097 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3641, A3642, A3643, A3644, A3645 |
|
A:A1301, A:A1635, A:A1637, G:A1299, G:A1300 |
|
|
|
4tpb |
A3097 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3642, A3643, A3644, A3645, A3646 |
|
A:A1301, A:A1635, A:A1637, G:A1299, G:A1300 |
|
|
|
4tpd |
A3097 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3652, A3653, A3654, A3655, A3656 |
|
A:A1301, A:A1635, A:A1637, G:A1299, G:A1300 |
|
|
|
4tp7 |
A3098 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3642, A3643, A3644, A3645, A3646 |
|
A:A1301, A:A1635, A:A1637, G:A1299, G:A1300 |
|
|
|
4tpf |
A3098 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3645, A3646, A3647, A3648, A3649 |
|
A:A1301, A:A1635, A:A1637, G:A1299, G:A1300 |
|
|
|
4tox |
A3098 |
Oph·5Pout |
U:A1636(OP2) |
|
|
A3646, A3647, A3648, A3649, A3650 |
|
A:A1301, A:A1635, A:A1637, G:A1299, G:A1300 |
|
|
|
3v2f |
A3235 |
Oph·5Pout |
C:A1636(OP2) |
|
|
A4047, A4048, A4049, A4050, A4051 |
|
G:A1299, U:A1300, G:A1635, A:A1301, A:A1637 |
|
|
|
3v25 |
A3286 |
Oph·5Pout |
C:A1636(OP2) |
|
|
A3698, A3699, A3700, A3701, A3702 |
|
G:A1299, U:A1300, G:A1635, A:A1301, A:A1637 |
|
|
|
4nw1 |
A3551 |
Oph·5Pout |
C:A1636(OP2) |
|
|
A4710, A4711, A4712, A4713 |
|
G:A1299, U:A1300, A:A1301, G:A1635, A:A1637 |
|
|
|
4qcx |
A3555 |
Oph·5Pout |
C:A1636(OP2) |
|
|
A4628, A4629, A4630, A4631 |
|
G:A1299, A:A1301, U:A1300, G:A1635, A:A1637 |
|
|
|
4nvx |
A3557 |
Oph·5Pout |
C:A1636(OP2) |
|
|
A4711, A4712, A4713, A4714 |
|
G:A1299, U:A1300, G:A1635, A:A1301, A:A1637 |
|
|
|
1yjw |
78035 |
Oph·5Pout |
G:01484(OP2) |
|
|
07233, 07235, 07237, 07289, 07290 |
|
C:01455, C:01456, U:01457, A:01485, C:01483 |
|
U:01457, G:01484 |
|
1nji |
A8060 |
Oph·5Pout |
G:A175(OP2) |
|
|
A7818, A7819, A7820, A9167 |
|
C:A173, G:A225, U:A172, G:A393, U:A392 |
|
|
|
1jj2 |
08060 |
Oph·5Pout |
G:0175(OP2) |
|
|
07373, 07374, 07375, 08687 |
|
C:0173, G:0225, U:0172, G:0393, U:0392 |
|
|
|
1k9m |
A8060 |
Oph·5Pout |
G:A175(OP2) |
|
|
A7836, A7837, A7838, A9172 |
|
C:A173, G:A225, U:A172, G:A393, U:A392 |
|
|
|
2otj |
08060 |
Oph·5Pout |
G:0175(OP2) |
|
|
07820, 07821, 07822, 09172 |
|
C:0173, G:0225, U:0172, G:0393, U:0392 |
|
|
|
1s72 |
08060 |
Oph·5Pout |
G:0175(OP2) |
|
|
07328, 07329, 07330, 08687 |
|
C:0173, G:0225, U:0172, G:0393, U:0392 |
|
|
|
3cc2 |
08060 |
Oph·5Pout |
G:0175(OP2) |
|
|
07386, 07387, 07388, 08694 |
|
C:0173, G:0225, U:0172, G:0393, U:0392 |
|
|
|