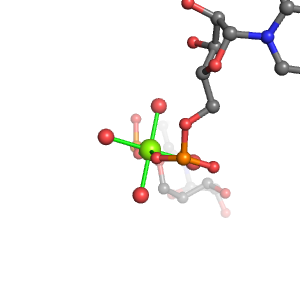
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3cc2 |
08077 |
trans-2Oph |
C:0880(OP1), U:0883(OP2) |
|
|
03176, 07404, 07405, 08769 |
|
A:0882, C:0884 |
|
U:0883, A:01836 |
|
3cc2 |
08015 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03175, 07350, 08527, 08545 |
|
|
G:01688 |
A:0844, A:01689, C:01690, A:0882, C:01692 |
|
3cc2 |
08052 |
trans-2Oph |
U:01125(OP1), G:992(OP1) |
|
|
03177, 07452, 98390, 98416 |
|
C:01126 |
|
C:01129, G:990 |
|
3cc7 |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03655, 07838, 09012, 09030 |
|
|
G:01688 |
A:0844, A:01689, C:01690, A:0882, C:01692 |
|
3cce |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03643, 07813, 09029, 18953 |
|
|
G:01688 |
A:0844, A:01689, C:01690, C:01692, A:0882 |
|
3ccj |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03617, 07701, 09012, 09032 |
|
|
G:01688 |
A:0844, A:01689, C:01690, A:0882, C:01692 |
|
3ccl |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03646, 07817, 09031, 18953 |
|
|
G:01688 |
A:0844, A:01689, C:01690, C:01692, A:0882 |
|
3ccm |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03650, 07822, 09013, 09031 |
|
|
G:01688 |
A:0844, A:0882, A:01689, C:01690, C:01692 |
|
3ccq |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03643, 07838, 09013, 09032 |
|
|
G:01688 |
A:0844, A:01689, C:01690, A:0882, C:01692 |
|
3ccr |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03628, 07783, 09012, 09032 |
|
|
G:01688 |
A:0844, A:01689, C:01690, A:0882, C:01692 |
|
3ccs |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03622, 07792, 09013, 09030 |
|
|
G:01688 |
A:0844, A:01689, C:01690, A:0882, C:01692 |
|
3ccu |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03636, 07822, 09028, 18953 |
|
|
G:01688 |
A:0844, A:01689, C:01690, C:01692, A:0882 |
|
3ccv |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03630, 07797, 09027, 18953 |
|
|
G:01688 |
A:0844, A:01689, C:01690, C:01692, A:0882 |
|
3cd6 |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03649, 07833, 09030, 18953 |
|
|
G:01688 |
A:0844, A:01689, C:01690, C:01692, A:0882 |
|
3cpw |
08011 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
04061, 08218, 09520, Z8953 |
|
|
G:01688 |
A:0844, A:01689, C:01690, C:01692, A:0882 |
|
3cxc |
08015 |
trans-2Oph |
A:0844(OP1), A:01689(OP1) |
|
|
03004, 03021, 04570, 08683 |
|
|
G:01688 |
A:0882, C:01692, A:0844, A:01689, C:01690 |
|
3cxc |
08077 |
trans-2Oph |
C:0880(OP1), U:0883(OP2) |
|
|
03227, 04571, 08736, 08737 |
|
A:0882, C:0884 |
|
U:0883, A:01836 |
|
3cxc |
98052 |
trans-2Oph |
U:01125(OP1), G:93092(OP1) |
|
|
04572, 04806, 08783, 98390 |
|
C:01126 |
|
C:01129, G:93090 |
|
3df2 |
B3326 |
trans-2Oph |
A:B751(OP1), A:B1614(OP1) |
|
|
B4327, B4328, B4329, B4330 |
|
|
|
A:B1614, C:B1615, C:B1617, A:B789 |
|
3df4 |
B3087 |
trans-2Oph |
U:B999(OP2), C:B1153(OP2) |
|
|
B4088, B4089, B4090, B4091 |
|
C:B998 |
|
G:B1154 |
|
3df4 |
B3327 |
trans-2Oph |
A:B751(OP1), A:B1614(OP1) |
|
|
B4328, B4329, B4330, B4331 |
|
|
|
A:B1614, C:B1615, C:B1617, A:B751, A:B789 |
|
3df4 |
B3579 |
trans-2Oph |
C:B787(OP1), U:B790(OP2) |
|
|
B4580, B4581, B4582, B4583 |
|
C:B791 |
|
A:B1780 |
|
3hhn |
C1021 |
trans-2Oph |
C:C70(OP1), U:C106(OP1) |
|
|
C1037, C1038, C1039, C1040 |
|
G:C72 |
A:C71 |
|
|
3hhn |
E1021 |
trans-2Oph |
C:E70(OP1), U:E106(OP1) |
|
|
E1039, E1040, E1041, E1042 |
|
G:E72 |
A:E71 |
|
|
3i1n |
A2994 |
trans-2Oph |
A:A1359(OP1), G:A1360(OP2) |
|
|
A3463, A3464, A3465, A3466 |
|
|
|
G:A1361 |
|