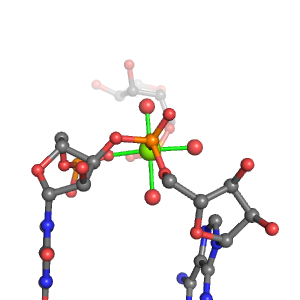
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1vs8 |
B2990 |
mer-3Oph |
C:B740(OP2), A:B1783(OP1), A:B1784(OP2) |
|
|
B3423, B3424, B3425 |
|
U:B741 |
|
|
|
1vs8 |
B2988 |
mer-3Oph |
A:B1780(OP1), U:B1782(OP1), A:B1783(OP2) |
|
|
B3417, B3418, B3419 |
|
|
|
A:B1783 |
|
1vt2 |
A2935 |
mer-3Oph |
U:A827(OP1), G:A2429(OP2), A:A2430(OP2) |
|
|
A3190, A3191, A3192 |
|
|
|
|
|
1w2b |
03919 |
mer-3Oph |
A:01836(OP1), U:01838(OP1), A:01839(OP2) |
|
|
07079, 07085, 07088 |
|
G:01837 |
|
A:01839 |
|
1yhq |
08005 |
mer-3Oph |
A:01836(OP1), U:01838(OP1), A:01839(OP2) |
|
|
04108, 08294, 08295 |
|
G:01837 |
|
A:01839 |
|
1yhq |
08006 |
mer-3Oph |
U:0919(OP1), C:02464(OP1), A:02465(OP1) |
|
|
04125, 08287, 08367 |
|
G:02466 |
|
|
|
1yi2 |
08007 |
mer-3Oph |
U:0832(OP2), A:01839(OP1), A:01840(OP2) |
|
|
03358, 07727, 09006 |
|
G:0833 |
|
|
|
1yi2 |
08008 |
mer-3Oph |
U:0919(OP1), C:02464(OP1), A:02465(OP1) |
|
|
03627, 07731, 07814 |
|
G:02466, C:0920 |
|
|
|
1yi2 |
08081 |
mer-3Oph |
C:01420(OP1), C:01421(OP2), G:01438(OP1) |
|
|
04094, 09613 |
|
|
|
|
|
1yi2 |
08005 |
mer-3Oph |
A:01836(OP1), U:01838(OP1), A:01839(OP2) |
|
|
03609, 07738, 07739 |
|
G:01837 |
|
A:01839 |
|
1yij |
08008 |
mer-3Oph |
U:0919(OP1), C:02464(OP1), A:02465(OP1) |
|
|
03639, 07743, 07827 |
|
G:02466, C:0920 |
|
|
|
1yij |
08005 |
mer-3Oph |
A:01836(OP1), U:01838(OP1), A:01839(OP2) |
|
|
03621, 07750, 07751 |
|
G:01837 |
|
A:01839 |
|
1yij |
08007 |
mer-3Oph |
U:0832(OP2), A:01839(OP1), A:01840(OP2) |
|
|
03367, 07739, 09006 |
|
G:0833 |
|
|
|
1yij |
08081 |
mer-3Oph |
C:01420(OP1), C:01421(OP2), G:01438(OP1) |
|
|
04112, 09615 |
|
|
|
|
|
1yj9 |
08007 |
mer-3Oph |
U:0832(OP2), A:01839(OP1), A:01840(OP2) |
|
|
03171, 07552, 08828 |
|
G:0833 |
|
|
|
1yj9 |
08008 |
mer-3Oph |
U:0919(OP1), C:02464(OP1), A:02465(OP1) |
|
|
03444, 07556, 07637 |
|
G:02466, C:0920 |
|
|
|
1yj9 |
08005 |
mer-3Oph |
A:01836(OP1), U:01838(OP1), A:01839(OP2) |
|
|
03426, 07563, 07564 |
|
G:01837 |
|
A:01839 |
|
1yj9 |
08081 |
mer-3Oph |
C:01420(OP1), C:01421(OP2), G:01438(OP1) |
|
|
03919, 09431 |
|
U:01422 |
|
|
|
1yjn |
08007 |
mer-3Oph |
U:0832(OP2), A:01839(OP1), A:01840(OP2) |
|
|
03366, 07751, 09006 |
|
G:0833 |
|
|
|
1yjn |
08008 |
mer-3Oph |
U:0919(OP1), C:02464(OP1), A:02465(OP1) |
|
|
03641, 07755, 07840 |
|
G:02466 |
|
|
|
1yjn |
08005 |
mer-3Oph |
A:01836(OP1), U:01838(OP1), A:01839(OP2) |
|
|
03622, 07762, 07763 |
|
G:01837 |
|
A:01839 |
|
1yjn |
08081 |
mer-3Oph |
C:01420(OP1), C:01421(OP2), G:01438(OP1) |
|
|
04114, 09614 |
|
|
|
|
|
1yjw |
78008 |
mer-3Oph |
U:0919(OP1), C:02464(OP1), A:02465(OP1) |
|
|
06154, 06155, 09018 |
|
G:02466 |
|
|
|
1yjw |
78005 |
mer-3Oph |
A:01836(OP1), U:01838(OP1), A:01839(OP2) |
|
|
07931, 07933, 07942 |
|
G:01837 |
|
A:01839 |
|
2avy |
A1594 |
mer-3Oph |
A:A116(OP2), G:A117(OP2), G:A289(OP2) |
|
|
A1849, A1850, A1851 |
|
A:A288 |
|
G:A117, U:A118 |
|