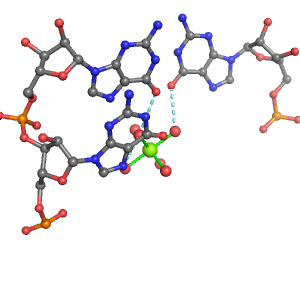
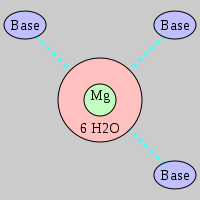
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4gau |
A3088 |
3Bout |
|
|
|
A3620, A3621, A3622, A3623, A3624, A3625 |
|
|
|
U:A1411, A:A1590, G:A1410 |
|
4tpd |
A3092 |
3Bout |
|
|
|
A3624, A3625, A3626, A3627, A3628, A3629 |
|
|
|
U:A1411, U:A1412, G:A1410 |
|
4tp5 |
A3092 |
3Bout |
|
|
|
A3623, A3624, A3625, A3626, A3627, A3628 |
|
|
|
U:A1411, G:A1410, A:A1591 |
|
4tpf |
A3093 |
3Bout |
|
|
|
A3617, A3618, A3619, A3620, A3621, A3622 |
|
|
|
G:A1410, U:A1411, U:A1409 |
|
4tp7 |
A3093 |
3Bout |
|
|
|
A3614, A3615, A3616, A3617, A3618, A3619 |
|
|
|
G:A1410, U:A1411, U:A1409 |
|
4tp9 |
A3093 |
3Bout |
|
|
|
A3621, A3622, A3623, A3624, A3625, A3626 |
|
|
|
U:A1411, U:A1412, G:A1410 |
|
4tom |
A3094 |
3Bout |
|
|
|
A3636, A3637, A3638, A3639, A3640, A3641 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
4tp9 |
A3096 |
3Bout |
|
|
|
A3637, A3638, A3639, A3640, A3641, A3642 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
4tp1 |
A3096 |
3Bout |
|
|
|
A3638, A3639, A3640, A3641, A3642, A3643 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
4tox |
A3096 |
3Bout |
|
|
|
A3634, A3635, A3636, A3637, A3638, A3639 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
4tp1 |
A3100 |
3Bout |
|
|
|
A3658, A3659, A3660, A3661, A3662, A3663 |
|
|
|
G:A1823, G:A1824, U:A1796 |
|
4too |
A3103 |
3Bout |
|
|
|
A3666, A3667, A3668, A3669, A3670, A3671 |
|
|
|
C:A2047, G:A2049, G:A2618 |
|
4tox |
A3104 |
3Bout |
|
|
|
A3671, A3672, A3673, A3674, A3675, A3676 |
|
|
|
G:A2049, G:A2048, G:A2618 |
|
2qam |
B3117 |
3Bout |
|
|
|
B3706, B3707, B3708, B3709, B3710, B3711 |
|
|
|
A:B756, A:B739, G:B757 |
|
2qba |
B3117 |
3Bout |
|
|
|
B3706, B3707, B3708, B3709, B3710, B3711 |
|
|
|
A:B756, A:B739, G:B757 |
|
4qcu |
A3127 |
3Bout |
|
|
|
A4042, A4043, A4044, A4045, P101 |
|
|
|
G:A44, G:A396, C:A43 |
|
4tpf |
A3132 |
3Bout |
|
|
|
A3781, A3782, A3783, A3784, C307, C308 |
|
|
|
U:A1825, C:A1793, G:A1826 |
|
4tp7 |
A3132 |
3Bout |
|
|
|
A3778, A3779, A3780, A3781, C310, C311 |
|
|
|
U:A1825, C:A1793, G:A1826 |
|
4too |
A3132 |
3Bout |
|
|
|
A3777, A3778, A3779, A3780, A3781, A3782 |
|
|
|
U:A1825, G:A1826, U:A1827 |
|
4tox |
A3132 |
3Bout |
|
|
|
A3782, A3783, A3784, A3785, A3786, C307 |
|
|
|
C:A1793, U:A1825, G:A1826 |
|
4qd0 |
A3137 |
3Bout |
|
|
|
A4093, A4094, A4095, A4096, A4097 |
|
|
|
A:A684, G:A682, G:A683 |
|
4qcy |
A3147 |
3Bout |
|
|
|
A4041, A4042, A4043, A4044, A4045 |
|
|
|
G:A44, G:A396, C:A43 |
|
4nvu |
A3155 |
3Bout |
|
|
|
A4032, A4033, A4034, A4035 |
|
|
|
G:A44, U:A45, G:A396 |
|
4nvy |
A3155 |
3Bout |
|
|
|
A4033, A4034, A4035, P101 |
|
|
|
G:A44, U:A45, G:A396 |
|
3df2 |
B3240 |
3Bout |
|
|
|
B4241, B4242, B4243, B4244, B4245 |
|
|
|
G:B2303, U:B2302, G:B2304 |
|