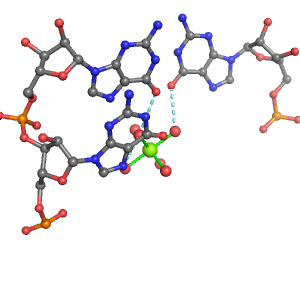
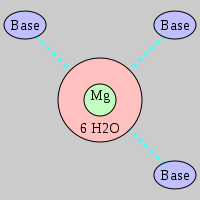
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3hhn |
C1025 |
3Bout |
|
|
|
C1057, C1058, C1059, C1060, C1061, C1062 |
|
|
|
G:C87, G:C88, A:C103 |
|
3hhn |
E1024 |
3Bout |
|
|
|
E66, E1052, E1053, E1054, E1055, E1056 |
|
|
|
G:E75, A:E80, G:E81 |
|
3hhn |
C60 |
3Bout |
|
|
|
C59, C61, C62, C63, C64, C65 |
|
|
|
G:C81, A:C80, G:C75 |
|
3i1n |
A2995 |
3Bout |
|
|
|
A3467, A3468, A3469, A3470, A3471, A3472 |
|
|
|
G:A1410, U:A1411, A:A1590 |
|
3i1n |
A2998 |
3Bout |
|
|
|
A3483, A3484, A3485, A3486, A3487, A3488 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
3i1n |
A2919 |
3Bout |
|
|
|
A3118, A3119, A3120, A3121, A3122, A3123 |
|
|
|
G:A1259, U:A580, U:A1258 |
|
3i1o |
A1558 |
3Bout |
|
|
|
A1708, A1709, A1710, A1711, A1712 |
|
|
|
G:A1371, U:A1372, G:A1370 |
|
3i1p |
A2972 |
3Bout |
|
|
|
A3359, A3360, A3361, A3362, A3363, A3364 |
|
|
|
G:A2242, U:A2243, A:A2241 |
|
3i1r |
A2904 |
3Bout |
|
|
|
A3040, A3041, A3042, A3043, A3044, A3045 |
|
|
|
G:A17, U:A521, U:A18 |
|
3i1r |
A2919 |
3Bout |
|
|
|
A3117, A3118, A3119, A3120, A3121, A3122 |
|
|
|
U:A580, U:A1258, G:A1259 |
|
3i1t |
B119 |
3Bout |
|
|
|
B627, B628, B629, B630, B631 |
|
|
|
U:B80, U:B95, G:B96 |
|
3i1t |
A2930 |
3Bout |
|
|
|
A3167, A3168, A3169, A3170, E271, E276 |
|
|
|
G:A797, G:A798, C:A679 |
|
3i1t |
A2982 |
3Bout |
|
|
|
A3411, A3412, A3413, A3414, A3415, A3416 |
|
|
|
G:A2549, G:A2550, C:A2551 |
|
3i1z |
A1557 |
3Bout |
|
|
|
A1686, A1687, A1688, A1689, A1690, A1691 |
|
|
|
G:A800, U:A798, G:A799 |
|
3i20 |
A3005 |
3Bout |
|
|
|
A3520, A3521, A3522, A3523, A3524, A3525 |
|
|
|
C:A2047, G:A2048, G:A2049 |
|
3i20 |
A2998 |
3Bout |
|
|
|
A3482, A3483, A3484, A3485, A3486, A3487 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
3i20 |
A2904 |
3Bout |
|
|
|
A3040, A3041, A3042, A3043, A3044, A3045 |
|
|
|
U:A521, G:A17, U:A18 |
|
3i20 |
A2924 |
3Bout |
|
|
|
A3141, A3142, A3143, A3144, A3145, A3146 |
|
|
|
A:A756, A:A739, G:A757 |
|
3i20 |
A2919 |
3Bout |
|
|
|
A3117, A3118, A3119, A3120, A3121, A3122 |
|
|
|
G:A1259, U:A580, U:A1258 |
|
3i20 |
A3001 |
3Bout |
|
|
|
A3502, A3503, A3504, A3505, A3506, A3507 |
|
|
|
G:A1823, G:A1824, U:A1825 |
|
3i22 |
A2925 |
3Bout |
|
|
|
A3146, A3147, A3148, A3149, A3150, A3151 |
|
|
|
G:A757, A:A739, A:A756 |
|
3i22 |
A2923 |
3Bout |
|
|
|
A3136, A3137, A3138, A3139, A3140, A3141 |
|
|
|
C:A650, G:A638, G:A651 |
|
3ivk |
C129 |
3Bout |
|
|
|
C171, C172, C173, C174, C175, C176 |
|
|
|
G:C17, G:C18, U:C16 |
|
3ivk |
M125 |
3Bout |
|
|
|
M156, M157, M158, M159, M160, M161 |
|
|
|
A:M80, G:M81, G:M75 |
|
3ivk |
M124 |
3Bout |
|
|
|
M150, M151, M152, M153, M154, M155 |
|
|
|
G:M72, A:M73, G:M105 |
|