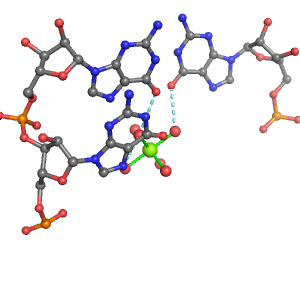
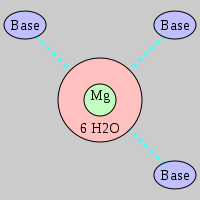
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3ivk |
C124 |
3Bout |
|
|
|
C150, C151, C152, C153, C154, C155 |
|
|
|
G:C72, A:C73, G:C105 |
|
3ivk |
C129 |
3Bout |
|
|
|
C171, C172, C173, C174, C175, C176 |
|
|
|
G:C17, G:C18, U:C16 |
|
3ivk |
M124 |
3Bout |
|
|
|
M150, M151, M152, M153, M154, M155 |
|
|
|
G:M72, A:M73, G:M105 |
|
3oaq |
A1570 |
3Bout |
|
|
|
A1747, A1748, A1749, A1750, A1751, U219 |
|
|
|
G:A1525, G:A1526, C:A1510 |
|
3oar |
A1560 |
3Bout |
|
|
|
A1707, A1708, A1709, A1710, I169 |
|
|
|
G:A1371, U:A1372, G:A1370 |
|
3oas |
A2904 |
3Bout |
|
|
|
A3037, A3038, A3039, A3040, A3041, A3042 |
|
|
|
U:A18, U:A521, G:A17 |
|
3oat |
A2995 |
3Bout |
|
|
|
A3463, A3464, A3465, A3466, A3467, A3468 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
3ofc |
A2922 |
3Bout |
|
|
|
A3134, A3135, A3136, A3137, A3138, A3139 |
|
|
|
G:A757, A:A756, A:A739 |
|
3ofc |
A2996 |
3Bout |
|
|
|
A3476, A3477, A3478, A3479, A3480, A3481 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
3ofc |
A2993 |
3Bout |
|
|
|
A3460, A3461, A3462, A3463, A3464, A3465 |
|
|
|
U:A1411, G:A1410, A:A1590 |
|
3ofd |
A2905 |
3Bout |
|
|
|
A3038, A3039, A3040, A3041, A3042, A3043 |
|
|
|
U:A18, U:A521, G:A17 |
|
3ofd |
A2971 |
3Bout |
|
|
|
A3358, A3359, A3360, A3361, A3362, A3363 |
|
|
|
G:A2242, U:A2243, A:A2241 |
|
3ofo |
A1568 |
3Bout |
|
|
|
A1743, A1744, A1745, A1746, U216, U219 |
|
|
|
C:A1509, G:A1525, G:A1526 |
|
3ofq |
A2920 |
3Bout |
|
|
|
A3114, A3115, A3116, A3117, A3118, A3119 |
|
|
|
C:A581, G:A1259, U:A1258 |
|
3ofr |
A2997 |
3Bout |
|
|
|
A3473, A3474, A3475, A3476, A3477, A3478 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
3ofr |
A3035 |
3Bout |
|
|
|
A3624, A3625, C759, C760, C762 |
|
|
|
G:A2592, U:A2593, G:A2599 |
|
3ofz |
A2999 |
3Bout |
|
|
|
A3485, A3486, A3487, A3488, A3489, A3490 |
|
|
|
G:A1824, U:A1825, G:A1823 |
|
3ofz |
A2992 |
3Bout |
|
|
|
A3449, A3450, A3451, A3452, A3453, A3454 |
|
|
|
G:A1410, U:A1411, U:A1409 |
|
3ofz |
A2995 |
3Bout |
|
|
|
A3465, A3466, A3467, A3468, A3469, A3470 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
3og0 |
A2904 |
3Bout |
|
|
|
A3036, A3037, A3038, A3039, A3040, A3041 |
|
|
|
U:A18, U:A521, G:A17 |
|
3og0 |
A2918 |
3Bout |
|
|
|
A3112, A3113, A3114, A3115, A3116, A3117 |
|
|
|
U:A580, C:A581, U:A1258 |
|
3or9 |
A1543 |
3Bout |
|
|
|
A1621, A1622, A1623, A1624, A1625, A1626 |
|
|
|
G:A592, U:A593, G:A645 |
|
3orb |
A2996 |
3Bout |
|
|
|
A3475, A3476, A3477, A3478, A3479, A3480 |
|
|
|
G:A1465, U:A1466, G:A1464 |
|
3ox0 |
A110 |
3Bout |
|
|
|
A146, A147, A148, A149, A150, A151 |
|
|
|
G:A25, G:A26, U:A27 |
|
3oxm |
A112 |
3Bout |
|
|
|
A124, A125, A129, A144, A145, A146 |
|
|
|
U:A46, G:A47, G:A55 |
|