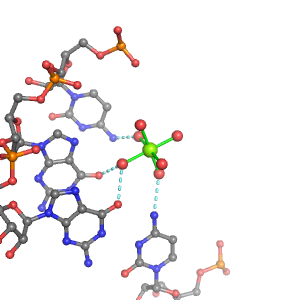
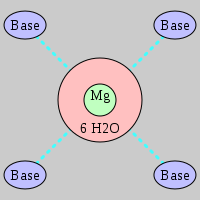
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1egk |
C13 |
4Bout |
|
|
|
C87, C88, C90, C91, C92, D89 |
|
|
|
U:C115, G:C116, DA:D126, DG:D124 |
|
1hr2 |
A20 |
4Bout |
|
|
|
A365, A366, A367, A368, A369, A370 |
|
|
|
U:A144, U:A156, U:A157, G:A158 |
|
3ivk |
C122 |
4Bout |
|
|
|
C138, C139, C140, C141, C142, C143 |
|
|
|
A:C97, G:C91, G:C92, U:C93 |
|
2g91 |
A191 |
4Bout |
|
|
|
A116, A120, A162, A180, A184, B122 |
|
|
|
G:A4, C:A5, U:A3, U:B12 |
|
2g91 |
B192 |
4Bout |
|
|
|
A139, A140, B138, B141 |
|
|
|
G:A2, U:A3, G:B15, C:B14 |
|
4a17 |
0203 |
4Bout |
|
|
|
07043, 07044, 07045, 07046, 07047, 07048 |
|
|
|
U:396, G:397, U:395, U:379 |
|
3v2f |
B210 |
4Bout |
|
|
|
B328, B329, B330, B331, B332 |
|
|
|
U:B80, G:B98, U:B96, G:B97 |
|
3v27 |
B222 |
4Bout |
|
|
|
B347, B348, B349, B350, B351, B352 |
|
|
|
U:B80, U:B96, G:B97, G:B98 |
|
1yls |
B243 |
4Bout |
|
|
|
A361, A366, B362, B363, B364, B365 |
|
|
|
G:A108, G:B202, G:B203, G:B201 |
|
1ehz |
A510 |
4Bout |
|
|
|
A712, A713, A714, A715, A716 |
|
|
|
G:A3, G:A4, U:A68, U:A69 |
|
2nug |
F513 |
4Bout |
|
|
|
E522, F542, F543, F544, F545, F552 |
|
|
|
G:E26, G:F26, U:E25, U:F25 |
|
3i1z |
A1543 |
4Bout |
|
|
|
A1621, A1622, A1623, A1624, A1625, A1626 |
|
|
|
G:A646, G:A592, U:A593, G:A645 |
|
3ofa |
A1543 |
4Bout |
|
|
|
A1620, A1621, A1622, A1623, A1624, A1625 |
|
|
|
G:A646, G:A592, U:A593, G:A645 |
|
3ofo |
A1543 |
4Bout |
|
|
|
A1619, A1620, A1621, A1622, A1623, A1624 |
|
|
|
G:A646, G:A592, U:A593, G:A645 |
|
2qp0 |
A1590 |
4Bout |
|
|
|
A2113, A2114, A2115, A2116, A2117 |
|
|
|
G:A1048, G:A1050, G:A1207, C:A1208 |
|
4tol |
A1610 |
4Bout |
|
|
|
A1744, A1745, A1746, A1747, A1748, A1749 |
|
|
|
G:A646, U:A593, U:A594, G:A645 |
|
4tp8 |
A1610 |
4Bout |
|
|
|
A1743, A1744, A1745, A1746, A1747, A1748 |
|
|
|
G:A646, U:A593, U:A594, G:A645 |
|
4gd2 |
A1620 |
4Bout |
|
|
|
A1792, A1793, A1794, A1795, A1796, A1797 |
|
|
|
G:A645, G:A592, U:A593, G:A646 |
|
4gd1 |
A1636 |
4Bout |
|
|
|
A1867, A1868, A1869, A1870, A1871, U101 |
|
|
|
G:A1525, C:A1524, G:A1526, C:A1509 |
|
4ji8 |
A1680 |
4Bout |
|
|
|
A2138, N201, N202, N203 |
|
|
|
A:A1360, G:A1361, C:A980, C:A979 |
|
4ji5 |
A1714 |
4Bout |
|
|
|
A2010, A2011, A2012, A2013, A2014, A2015 |
|
|
|
G:A44, U:A45, G:A396, C:A395 |
|
4ji1 |
A1723 |
4Bout |
|
|
|
A2028, A2029, A2030, A2031, A2032, A2033 |
|
|
|
G:A854, U:A833, G:A852, G:A853 |
|
4ji1 |
A1737 |
4Bout |
|
|
|
A2111, A2112, A2113, A2114, A2115, A2116 |
|
|
|
G:A494, G:A492, G:A490, G:A491 |
|
4dv1 |
A1764 |
4Bout |
|
|
|
A2091, A2092, A2093, A2094, A2095, A2096 |
|
|
|
G:A664, G:A741, G:A742, G:A666 |
|
4dv0 |
A1780 |
4Bout |
|
|
|
A2103, A2104, A2105, A2106 |
|
|
|
G:A664, G:A666, U:A740, G:A741 |
|