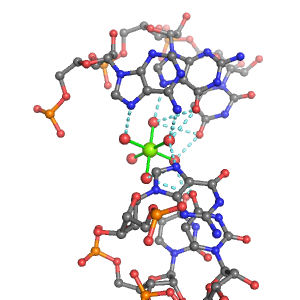
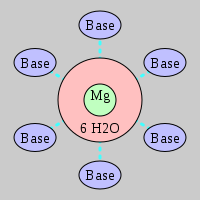
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2i2p |
A1548 |
6Bout |
|
|
|
A1628, A1629, A1630, A1631, A1632, A1633 |
|
|
|
G:A645, G:A646, G:A592, U:A644, U:A593, U:A594 |
|
4tp4 |
A1610 |
6Bout |
|
|
|
A1744, A1745, A1746, A1747, A1748, A1749 |
|
|
|
G:A646, U:A593, U:A594, G:A645, G:A592, U:A644 |
|
2qan |
A2114 |
6Bout |
|
|
|
A2115, A2116, A2117, A2118, A2119, A2120 |
|
|
|
U:A594, G:A645, G:A592, U:A593, G:A646, C:A647 |
|
2qox |
B2905 |
6Bout |
|
|
|
B3015, B3016, B3017, B3018, B3019, B3020 |
|
|
|
U:B2075, G:B2238, G:B2242, U:B2243, A:B2241, U:B2074 |
|
2awb |
B2905 |
6Bout |
|
|
|
B3015, B3016, B3017, B3018, B3019, B3020 |
|
|
|
G:B2238, G:B2242, U:B2243, A:B2241, U:B2074, U:B2075 |
|
2qbk |
B2905 |
6Bout |
|
|
|
B3017, B3018, B3019, B3020, B3021, B3022 |
|
|
|
G:B2238, G:B2242, A:B2241, U:B2074, U:B2075, U:B2243 |
|
1vs8 |
B2905 |
6Bout |
|
|
|
B3014, B3015, B3016, B3017, B3018, B3019 |
|
|
|
G:B2238, G:B2242, U:B2243, A:B2241, U:B2074, U:B2075 |
|
2i2t |
B2906 |
6Bout |
|
|
|
B3028, B3029, B3030, B3031, B3032, B3033 |
|
|
|
G:B2238, A:B2241, G:B2242, U:B2075, A:B2077, U:B2243 |
|
2qbi |
B2906 |
6Bout |
|
|
|
B3021, B3022, B3023, B3024, B3025, B3026 |
|
|
|
U:B2074, U:B2243, A:B2241, G:B2242, U:B2075, G:B2238 |
|
2z4n |
B2906 |
6Bout |
|
|
|
B3017, B3018, B3019, B3020, B3021, B3022 |
|
|
|
U:B2075, G:B2238, G:B2242, U:B2243, A:B2241, U:B2074 |
|
2qov |
B2906 |
6Bout |
|
|
|
B3020, B3021, B3022, B3023, B3024, B3025 |
|
|
|
U:B2074, U:B2243, A:B2241, U:B2075, G:B2238, G:B2242 |
|
2z4l |
B2907 |
6Bout |
|
|
|
B3021, B3022, B3023, B3024, B3025, B3026 |
|
|
|
U:B2074, U:B2243, A:B2241, G:B2242, U:B2075, G:B2238 |
|
3ofz |
A2967 |
6Bout |
|
|
|
A3334, A3335, A3336, A3337, A3338, A3339 |
|
|
|
U:A2075, G:A2238, G:A2242, U:A2074, U:A2243, A:A2241 |
|
3orb |
A2968 |
6Bout |
|
|
|
A3342, A3343, A3344, A3345, A3346, A3347 |
|
|
|
G:A2238, G:A2242, U:A2075, U:A2074, U:A2243, A:A2241 |
|
3i1n |
A2969 |
6Bout |
|
|
|
A3347, A3348, A3349, A3350, A3351, A3352 |
|
|
|
G:A2238, G:A2242, U:A2075, U:A2074, U:A2243, A:A2241 |
|
3i20 |
A2969 |
6Bout |
|
|
|
A3345, A3346, A3347, A3348, A3349, A3350 |
|
|
|
G:A2238, G:A2242, U:A2075, U:A2074, U:A2243, A:A2241 |
|
3ofr |
A2969 |
6Bout |
|
|
|
A3342, A3343, A3344, A3345, A3346, A3347 |
|
|
|
G:A2238, G:A2242, U:A2075, U:A2074, U:A2243, A:A2241 |
|
3i1r |
A2969 |
6Bout |
|
|
|
A3345, A3346, A3347, A3348, A3349, A3350 |
|
|
|
G:A2238, G:A2242, U:A2075, U:A2074, U:A2243, A:A2241 |
|
2qbc |
B3001 |
6Bout |
|
|
|
B3620, B3621, B3622, B3623, B3624, B3625 |
|
|
|
G:B2238, G:B2242, A:B2241, U:B2074, U:B2075, U:B2243 |
|
2qba |
B3007 |
6Bout |
|
|
|
B3619, B3620, B3621, B3622, B3623, B3624 |
|
|
|
U:B2074, U:B2243, A:B2241, G:B2242, U:B2075, G:B2238 |
|
2qbe |
B3007 |
6Bout |
|
|
|
B3618, B3619, B3620, B3621, B3622, B3623 |
|
|
|
U:B2243, A:B2241, U:B2074, U:B2075, G:B2238, G:B2242 |
|
2qam |
B3007 |
6Bout |
|
|
|
B3618, B3619, B3620, B3621, B3622, B3623 |
|
|
|
U:B2074, U:B2243, A:B2241, G:B2242, U:B2075, G:B2238 |
|
4gar |
A3064 |
6Bout |
|
|
|
A3502, A3503, A3504, A3505, A3506, A3507 |
|
|
|
G:A2242, G:A2238, A:A2241, U:A2074, U:A2075, U:A2243 |
|
4peb |
A3065 |
6Bout |
|
|
|
A3503, A3504, A3505, A3506, A3507, A3508 |
|
|
|
G:A2238, A:A2241, G:A2242, U:A2075, U:A2074, U:A2243 |
|
4tom |
A3065 |
6Bout |
|
|
|
A3500, A3501, A3502, A3503, A3504, A3505 |
|
|
|
G:A2238, G:A2242, U:A2075, U:A2074, U:A2243, A:A2241 |
|