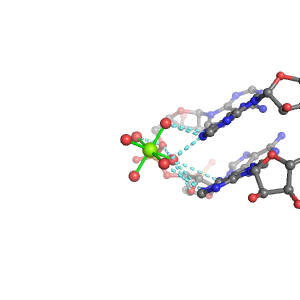
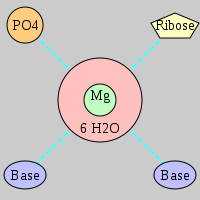
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1kqs |
08076 |
Pout·Rout·2Bout |
|
|
|
06351, 07324, 07325, 07377, 07378, 17875 |
|
U:01419 |
U:01418 |
G:01443, G:01444 |
|
1nji |
A8076 |
Pout·Rout·2Bout |
|
|
|
A6848, A7834, A7835, A7891, A7892, 37875 |
|
U:A1419 |
U:A1418 |
G:A1443, G:A1444 |
|
1n8r |
A8076 |
Pout·Rout·2Bout |
|
|
|
A7233, A8396, A8397, A8449, A8450, 37875 |
|
U:A1419 |
U:A1418 |
G:A1443, G:A1444 |
|
1m1k |
A8076 |
Pout·Rout·2Bout |
|
|
|
A6449, A7430, A7431, A7482, A7483, 37875 |
|
U:A1419 |
U:A1418 |
G:A1443, G:A1444 |
|
1jj2 |
08076 |
Pout·Rout·2Bout |
|
|
|
06397, 07389, 07390, 07445, 07446, 17875 |
|
U:01419 |
U:01418 |
G:01443, G:01444 |
|
1m90 |
A8076 |
Pout·Rout·2Bout |
|
|
|
A6391, A7372, A7373, A7427, A7428, 37875 |
|
U:A1419 |
U:A1418 |
G:A1443, G:A1444 |
|
1kc8 |
A8076 |
Pout·Rout·2Bout |
|
|
|
A6849, A7841, A7842, A7895, A7896, 37875 |
|
U:A1419 |
U:A1418 |
G:A1443, G:A1444 |
|
3cxc |
08076 |
Pout·Rout·2Bout |
|
|
|
07749, 08734, 08735, 08787, 08788, 17875 |
|
U:01419 |
U:01418 |
G:01443, G:01444 |
|
1vq7 |
08076 |
Pout·Rout·2Bout |
|
|
|
07100, 08154, 08155, 08207, 08208, 27875 |
|
U:01419 |
U:01418 |
G:01443, G:01444 |
|
1vq6 |
08076 |
Pout·Rout·2Bout |
|
|
|
07065, 08123, 08124, 08173, 08174, 08175 |
|
U:01419 |
U:01418 |
G:01443, G:01444 |
|
1q82 |
A8076 |
Pout·Rout·2Bout |
|
|
|
A6336, A7314, A7366, A7367, 37875 |
|
U:A1419 |
U:A1418 |
G:A1444, G:A1443 |
|
4bpe |
A5078 |
Pout·Rout·2Bout |
|
|
|
A2391, A2392, A2398, A2399, A2400, A2470 |
|
U:A1277 |
G:A1276 |
G:A1296, A:A1297 |
|
4bpo |
A5078 |
Pout·Rout·2Bout |
|
|
|
A2251, A2254, A2258, A2259, A2260, A2467 |
|
U:A1277 |
G:A1276 |
G:A1296, A:A1297 |
|
4bpn |
A5072 |
Pout·Rout·2Bout |
|
|
|
A6570, A6571, A6572, A6573, A6574, A6576 |
|
U:A1467 |
C:A1466 |
A:A1457, U:A1492 |
|
4bpe |
A5056 |
Pout·Rout·2Bout |
|
|
|
A2003, A2209, A2210, A2331, A2332, A2333 |
|
G:A1113 |
U:A8 |
G:A1113, G:A1114 |
|
4bpo |
A5056 |
Pout·Rout·2Bout |
|
|
|
A2002, A2214, A2215, A2426, A2427, A2428 |
|
G:A1113 |
U:A8 |
G:A1113, G:A1114 |
|
3v2d |
A3664 |
Pout·Rout·2Bout |
|
|
|
A5557, A5558, A5559, A5560, A5561 |
|
A:A983 |
U:A562 |
A:A572, C:A975 |
|
4qct |
A3619 |
Pout·Rout·2Bout |
|
|
|
A4920, A4921, A4922, A4923, A4924, A4925 |
|
G:A396 |
U:A395 |
G:A397, G:A398 |
|
3v27 |
A3608 |
Pout·Rout·2Bout |
|
|
|
A5108, A5109, A5110, A5111, A5112, A5113 |
|
A:A1142 |
U:A1142 |
G:A1016, G:A1017 |
|
3df4 |
B3602 |
Pout·Rout·2Bout |
|
|
|
B4603, B4604, B4605, B4606, B4607, B4608 |
|
A:B973 |
A:B972 |
A:B820, A:B821 |
|
3v23 |
A3589 |
Pout·Rout·2Bout |
|
|
|
A4928, A4929, A4930, A4931, A4932, A4933 |
|
A:A1142 |
U:A1142 |
G:A1017, G:A1016 |
|
3v27 |
A3561 |
Pout·Rout·2Bout |
|
|
|
A4921, A4922, A4923, A4924, A4925, A4926 |
|
U:A504 |
A:A503 |
G:A498, A:A482 |
|
3v27 |
A3518 |
Pout·Rout·2Bout |
|
|
|
A4718, A4719, A4720, A4721, A4722, A4723 |
|
A:A1528 |
G:A1527 |
U:A1540, G:A1541 |
|
3v29 |
A3426 |
Pout·Rout·2Bout |
|
|
|
A4126, A4127, A4128, A4129 |
|
U:A2696 |
G:A1707 |
G:A1758, G:A1707 |
|
4qcr |
A3341 |
Pout·Rout·2Bout |
|
|
|
A4165, A4166, A4167, A4168 |
|
U:A2708 |
G:A1754 |
G:A1789, G:A1754 |
|