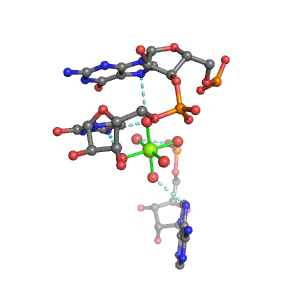
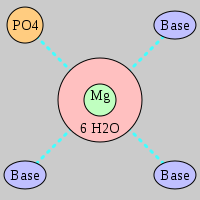
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1egk |
C9 |
Pout·3Bout |
|
|
|
B64, C64, C65, C67, C68, D63 |
|
G:C103 |
|
DG:B36, DA:B45, DG:D132 |
|
1egk |
A25 |
Pout·3Bout |
|
|
|
A46, A47, A48, B65, B66, D70 |
|
G:A3 |
|
DG:B32, DG:D136, DA:D135 |
|
1ffy |
T1202 |
Pout·3Bout |
|
|
|
T1298, T1299, T1300, T1301, T1302, T1303 |
|
A:T58 |
|
G:T53, U:T54, U:T55 |
|
1hr2 |
A16 |
Pout·3Bout |
|
|
|
A353, A354, A355, A356, A357, A358 |
|
A:A114 |
|
G:A116, U:A205, A:A115 |
|
1qu2 |
T1202 |
Pout·3Bout |
|
|
|
T1298, T1299, T1300, T1301, T1302, T1303 |
|
A:T58 |
|
G:T53, U:T54, U:T55 |
|
1vs5 |
A1563 |
Pout·3Bout |
|
|
|
A1696, A1697, A1698, A1699, A1700, A1701 |
|
G:A266 |
|
G:A257, G:A258, G:A259 |
|
1vs5 |
A1558 |
Pout·3Bout |
|
|
|
A1672, A1673, A1674, A1675, N142, N143 |
|
A:A1360 |
|
C:A980, G:A1361, A:A1360 |
|
1vs7 |
A1568 |
Pout·3Bout |
|
|
|
A1724, A1725, A1726, A1727, A1728, A1729 |
|
A:A781 |
|
U:A798, G:A800, G:A799 |
|
1vs7 |
A1567 |
Pout·3Bout |
|
|
|
A1718, A1719, A1720, A1721, A1722, A1723 |
|
G:A266 |
|
G:A257, G:A258, G:A259 |
|
1vs7 |
A1559 |
Pout·3Bout |
|
|
|
A1677, A1678, A1679, A1680, N150 |
|
A:A1360 |
|
C:A980, G:A1361, A:A1360 |
|
1vs8 |
B2956 |
Pout·3Bout |
|
|
|
B3263, B3264, B3265, B3266, B3267, B3268 |
|
A:B2386 |
|
C:B2326, U:B2387, A:B2386 |
|
1vt2 |
A2992 |
Pout·3Bout |
|
|
|
A3457, A3458, A3459, A3460, A3461, A3462 |
|
G:A977 |
|
G:A977, G:A978, G:A976 |
|
1vt2 |
A3015 |
Pout·3Bout |
|
|
|
A3547, A3548, A3549, A3550, A3551, 2647 |
|
U:A686 |
|
G:A771, U:A769, G:A770 |
|
1yj9 |
08071 |
Pout·3Bout |
|
|
|
03935, 05862, 07216, 07347, 07618 |
|
A:01815 |
|
U:01813, C:01818, U:01817 |
|
2avy |
A1557 |
Pout·3Bout |
|
|
|
A1671, A1672, N130, N131, N132, N135 |
|
A:A1360 |
|
C:A980, G:A1361, A:A1360 |
|
2avy |
A1563 |
Pout·3Bout |
|
|
|
A1699, A1700, A1701, A1702, A1703, A1704 |
|
A:A781 |
|
G:A799, G:A800, U:A798 |
|
2aw4 |
B2922 |
Pout·3Bout |
|
|
|
B3095, B3096, B3097, B3098, B3099, B3100 |
|
U:B1267 |
|
G:B2010, U:B2011, G:B2012 |
|
2aw7 |
A1558 |
Pout·3Bout |
|
|
|
A1678, A1679, A1680, A1681, N141 |
|
A:A1360 |
|
C:A980, A:A1360, G:A1361 |
|
2aw7 |
A1567 |
Pout·3Bout |
|
|
|
A1725, A1726, A1727, A1728, A1729, A1730 |
|
A:A781 |
|
G:A800, U:A798, G:A799 |
|
2aw7 |
A1566 |
Pout·3Bout |
|
|
|
A1719, A1720, A1721, A1722, A1723, A1724 |
|
G:A266 |
|
G:A257, G:A258, G:A259 |
|
2awb |
B2957 |
Pout·3Bout |
|
|
|
B3267, B3268, B3269, B3270, B3271, B3272 |
|
A:B2386 |
|
C:B2326, U:B2387, A:B2386 |
|
2gcs |
B505 |
Pout·3Bout |
|
|
|
B211, B300, B330, B331, B332, B333 |
|
A:B129 |
|
U:B100, G:B102, G:B101 |
|
2h0x |
B505 |
Pout·3Bout |
|
|
|
B211, B300, B330, B331, B332, B333 |
|
A:B129 |
|
U:B100, G:B102, G:B101 |
|
2i2p |
A1563 |
Pout·3Bout |
|
|
|
A1703, A1704, A1705, A1706, K165, K168 |
|
A:A781 |
|
G:A799, G:A800, U:A798 |
|
2i2p |
A1575 |
Pout·3Bout |
|
|
|
A1765, A1766, A1767, A1768, A1769 |
|
U:A1348 |
|
G:A1370, G:A1371, U:A1372 |
|