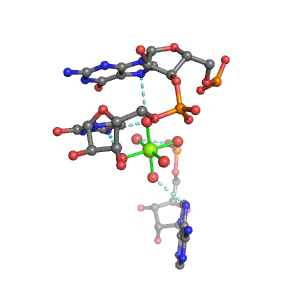
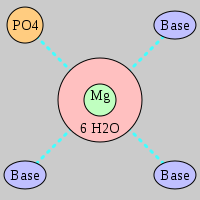
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1egk |
C9 |
Pout·3Bout |
|
|
|
B64, C64, C65, C67, C68, D63 |
|
G:C103 |
|
DG:B36, DA:B45, DG:D132 |
|
1hr2 |
A16 |
Pout·3Bout |
|
|
|
A353, A354, A355, A356, A357, A358 |
|
A:A114 |
|
G:A116, U:A205, A:A115 |
|
1egk |
A25 |
Pout·3Bout |
|
|
|
A46, A47, A48, B65, B66, D70 |
|
G:A3 |
|
DG:B32, DG:D136, DA:D135 |
|
4a1a |
079 |
Pout·3Bout |
|
|
|
05603, 05604, 05605, 05606, 05607, 05608 |
|
A:23 |
|
A:24, A:25, A:26 |
|
4oji |
A101 |
Pout·3Bout |
|
|
|
A245, A246, A247, A248, A249, A250 |
|
A:A29 |
|
G:A22, G:A23, U:A24 |
|
3ivk |
M132 |
Pout·3Bout |
|
|
|
M176, M177, M178, M179, M180, M181 |
|
A:M-4 |
|
A:M-4, G:M-3, U:M-2 |
|
4a1e |
0204 |
Pout·3Bout |
|
|
|
06803, 06804, 06805, 06806, 06807, 06808 |
|
A:374 |
|
U:376, A:3101, A:3100 |
|
4a17 |
0204 |
Pout·3Bout |
|
|
|
07059, 07060, 07061, 07062, 07063, 07064 |
|
A:374 |
|
U:376, A:3101, A:3100 |
|
3v27 |
B220 |
Pout·3Bout |
|
|
|
A4908, B339, B340, B341, B342, B343 |
|
C:A951 |
|
G:B85, G:B86, C:B92 |
|
2z75 |
B304 |
Pout·3Bout |
|
|
|
B411, B492, B493, B494, B544 |
|
A:B129 |
|
U:B100, G:B101, G:B102 |
|
4a1a |
0345 |
Pout·3Bout |
|
|
|
07955, 07956, 07957, 07958, 07959, 07960 |
|
A:289 |
|
U:270, G:271, G:290 |
|
2gcs |
B505 |
Pout·3Bout |
|
|
|
B211, B300, B330, B331, B332, B333 |
|
A:B129 |
|
U:B100, G:B102, G:B101 |
|
2h0x |
B505 |
Pout·3Bout |
|
|
|
B211, B300, B330, B331, B332, B333 |
|
A:B129 |
|
U:B100, G:B102, G:B101 |
|
1ffy |
T1202 |
Pout·3Bout |
|
|
|
T1298, T1299, T1300, T1301, T1302, T1303 |
|
A:T58 |
|
G:T53, U:T54, U:T55 |
|
1qu2 |
T1202 |
Pout·3Bout |
|
|
|
T1298, T1299, T1300, T1301, T1302, T1303 |
|
A:T58 |
|
G:T53, U:T54, U:T55 |
|
3i1s |
A1540 |
Pout·3Bout |
|
|
|
A1616, A1617, A1618, A1619, A1620, A1621 |
|
G:A645 |
|
G:A645, G:A592, U:A593 |
|
3ofp |
A1543 |
Pout·3Bout |
|
|
|
A1617, A1618, A1619, A1620, A1621, A1622 |
|
G:A645 |
|
G:A592, G:A646, G:A645 |
|
3ora |
A1543 |
Pout·3Bout |
|
|
|
A1621, A1622, A1623, A1624, A1625, A1626 |
|
G:A645 |
|
G:A646, G:A645, G:A592 |
|
3or9 |
A1544 |
Pout·3Bout |
|
|
|
A1627, A1628, A1629, A1630, A1631, A1632 |
|
G:A803 |
|
G:A771, U:A772, G:A773 |
|
3oaq |
A1544 |
Pout·3Bout |
|
|
|
A1627, A1628, A1629, A1630, A1631, A1632 |
|
G:A803 |
|
G:A771, U:A772, G:A773 |
|
2qou |
A1545 |
Pout·3Bout |
|
|
|
A1908, A1909, A1910, A1911, A1912, A1913 |
|
G:A803 |
|
U:A772, G:A773, G:A771 |
|
2i2u |
A1546 |
Pout·3Bout |
|
|
|
A1611, A1612, A1613, A1614, A1615, A1616 |
|
G:A803 |
|
U:A772, G:A773, G:A771 |
|
3i1o |
A1546 |
Pout·3Bout |
|
|
|
A1647, A1648, A1649, A1650, A1651, N122 |
|
A:A1360 |
|
C:A980, G:A1361, A:A1360 |
|
3or9 |
A1549 |
Pout·3Bout |
|
|
|
A1653, A1654, N101, N102, N103, N104 |
|
A:A1360 |
|
G:A1361, C:A980, A:A1360 |
|
3ora |
A1549 |
Pout·3Bout |
|
|
|
A1652, A1653, A1654, A1655, N101, N102 |
|
A:A1360 |
|
C:A980, G:A1361, A:A1360 |
|