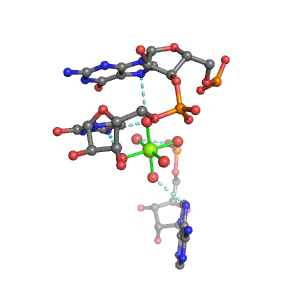
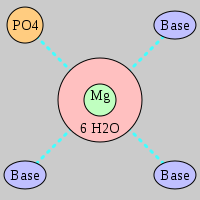
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3v26 |
A1735 |
Pout·3Bout |
|
|
|
A2058, A2059, A2060, A2061, A2062, A2063 |
|
G:A394 |
|
G:A44, U:A45, C:A395 |
|
3v27 |
A3650 |
Pout·3Bout |
|
|
|
A5337, A5338, A5339, A5340, A5341, A5342 |
|
A:A2765 |
|
G:A2766, G:A2736, G:A2737 |
|
3v27 |
B220 |
Pout·3Bout |
|
|
|
A4908, B339, B340, B341, B342, B343 |
|
C:A951 |
|
G:B85, G:B86, C:B92 |
|
3v27 |
A3617 |
Pout·3Bout |
|
|
|
A5159, A5160, A5161, A5162, A5163, A5164 |
|
G:A686 |
|
G:A771, A:A689, G:A690 |
|
3v27 |
A3355 |
Pout·3Bout |
|
|
|
A4050, A4051, A4052, A4053, A4054, 7103 |
|
A:A1308 |
|
G:A1311, G:A1309, G:A1310 |
|
3v27 |
A3565 |
Pout·3Bout |
|
|
|
A4944, A4945, A4946, A4947, A4948, A4949 |
|
A:A835 |
|
A:A835, G:A942, U:A943 |
|
3v27 |
A3611 |
Pout·3Bout |
|
|
|
A5123, A5124, A5125, A5126, A5127, A5128 |
|
G:A1855 |
|
G:A1856, G:A1857, G:A1855 |
|
3v27 |
A3647 |
Pout·3Bout |
|
|
|
A5319, A5320, A5321, A5322, A5323, A5324 |
|
G:A11 |
|
U:A12, G:A10, G:A11 |
|
3v27 |
A3448 |
Pout·3Bout |
|
|
|
A3968, A4420, A4421, A4422, A4423, A4424 |
|
A:A103 |
|
G:A82, G:A80, G:A81 |
|
3v27 |
A3626 |
Pout·3Bout |
|
|
|
A5213, A5214, A5215, A5216, A5217 |
|
U:A1267 |
|
G:A2010, U:A2011, G:A2009 |
|
3v28 |
A1714 |
Pout·3Bout |
|
|
|
A1889, A1890, A1891, A1892, A1893 |
|
A:A119 |
|
U:A287, G:A285, G:A286 |
|
3v29 |
A3538 |
Pout·3Bout |
|
|
|
A4541, A4542, A4543, A4544, A4545, A4546 |
|
G:A2193 |
|
G:A2193, G:A2194, C:A2095 |
|
3v29 |
A3404 |
Pout·3Bout |
|
|
|
A4037, A4038, A4039, A4040, A4041, A4042 |
|
A:A103 |
|
G:A80, G:A82, G:A81 |
|
3v29 |
A3501 |
Pout·3Bout |
|
|
|
A4396, A4397, A4398, A4399 |
|
A:A1701 |
|
G:A1699, G:A1702, G:A1703 |
|
3v2c |
A1802 |
Pout·3Bout |
|
|
|
A2306, A2307, A2308, A2309, A2310 |
|
A:A781 |
|
G:A799, G:A798, G:A800 |
|
3v2d |
A3692 |
Pout·3Bout |
|
|
|
A5657, A5658, A5659, A5660, A5661, A5662 |
|
U:A1267 |
|
G:A2010, U:A2011, G:A2009 |
|
3v2d |
A3675 |
Pout·3Bout |
|
|
|
A5590, A5591, A5592, A5593, A5594 |
|
G:A2845 |
|
G:A2845, G:A2846, G:A2848 |
|
3v2d |
A3633 |
Pout·3Bout |
|
|
|
A5463, A5464, A5465, A5466, A5467 |
|
A:A2765 |
|
G:A2735, G:A2736, G:A2737 |
|
3v2f |
A3603 |
Pout·3Bout |
|
|
|
A5085, A5086, A5087, A5088, A5089 |
|
C:A1398 |
|
G:A1400, G:A1385, G:A1401 |
|
3v2f |
A3345 |
Pout·3Bout |
|
|
|
A4402, A4403, A4404, A4405, A4406 |
|
A:A2360 |
|
C:A2356, U:A2357, A:A2361 |
|
3v2f |
A3367 |
Pout·3Bout |
|
|
|
A4472, A4473, A4474, A4475, A4476 |
|
A:A1308 |
|
G:A1309, G:A1310, G:A1311 |
|
3v2f |
A3600 |
Pout·3Bout |
|
|
|
A5071, A5072, A5073, A5074, A5075, A5076 |
|
U:A1267 |
|
G:A2010, G:A2009, U:A2011 |
|
3v2f |
A3612 |
Pout·3Bout |
|
|
|
A5117, A5118, A5119, A5120, A5121 |
|
G:A2845 |
|
G:A2848, G:A2845, G:A2846 |
|
4a17 |
0204 |
Pout·3Bout |
|
|
|
07059, 07060, 07061, 07062, 07063, 07064 |
|
A:374 |
|
U:376, A:3101, A:3100 |
|
4a1a |
079 |
Pout·3Bout |
|
|
|
05603, 05604, 05605, 05606, 05607, 05608 |
|
A:23 |
|
A:24, A:25, A:26 |
|