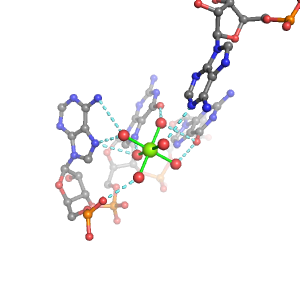
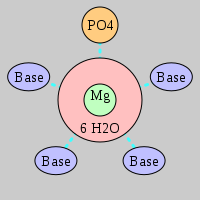
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1hr2 |
B17 |
Pout·4Bout |
|
|
|
B362, B363, B364, B365, B366, B367 |
|
A:B114 |
|
A:B115, G:B116, U:B205, G:B117 |
|
1vs5 |
A1549 |
Pout·4Bout |
|
|
|
A1629, A1630, A1631, A1632, A1633, A1634 |
|
U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
1vs8 |
B2923 |
Pout·4Bout |
|
|
|
B3105, B3106, B3107, B3108, B3109, B3110 |
|
U:B1267 |
|
U:B2011, G:B2012, G:B2010, A:B2009 |
|
1vt2 |
A2980 |
Pout·4Bout |
|
|
|
A3401, A3402, A3403, A3404, A3405, A3406 |
|
G:A2454 |
|
G:A2455, C:A2496, G:A2454, A:A2453 |
|
1vt2 |
A2943 |
Pout·4Bout |
|
|
|
A3224, A3225, A3226, A3227, A3228, A3229 |
|
U:A1267 |
|
A:A2009, G:A2010, U:A2011, G:A2012 |
|
2aw4 |
B2983 |
Pout·4Bout |
|
|
|
B3384, B3385, B3386, B3387, B3388, B3389 |
|
C:B2452 |
|
G:B2454, A:B2497, G:B2455, C:B2496 |
|
2awb |
B2923 |
Pout·4Bout |
|
|
|
B3106, B3107, B3108, B3109, B3110, B3111 |
|
U:B1267 |
|
U:B2011, G:B2012, G:B2010, A:B2009 |
|
2awb |
B2930 |
Pout·4Bout |
|
|
|
B3140, B3141, B3142, B3143, B3144, B3145 |
|
A:B2453 |
|
A:B2453, G:B2454, A:B2497, G:B2455 |
|
2i2p |
A1562 |
Pout·4Bout |
|
|
|
A1697, A1698, A1699, A1700, A1701, A1702 |
|
G:A266 |
|
G:A257, G:A259, G:A258, U:A268 |
|
2i2v |
B2930 |
Pout·4Bout |
|
|
|
B3150, B3151, B3152, B3153, B3154, B3155 |
|
A:B2453 |
|
A:B2453, G:B2454, A:B2497, G:B2455 |
|
2i2v |
B2923 |
Pout·4Bout |
|
|
|
B3115, B3116, B3117, B3118, B3119, B3120 |
|
U:B1267 |
|
U:B2011, G:B2012, G:B2010, A:B2009 |
|
2qal |
A2115 |
Pout·4Bout |
|
|
|
A2116, A2117, A2118, A2119, A2120, A2121 |
|
G:A266 |
|
G:A257, U:A268, G:A258, G:A259 |
|
2qal |
A2129 |
Pout·4Bout |
|
|
|
A2130, A2131, A2132, A2133, A2134, A2135 |
|
A:A559 |
|
U:A24, G:A9, A:A10, A:A559 |
|
2qam |
B3103 |
Pout·4Bout |
|
|
|
B3694, B3695, B3696, B3697, B3698, B3699 |
|
U:B1267 |
|
G:B2010, U:B2011, G:B2012, A:B2009 |
|
2qan |
A2141 |
Pout·4Bout |
|
|
|
A2142, A2143, A2144, A2145, A2146, A2147 |
|
G:A266 |
|
G:A257, G:A258, U:A268, G:A259 |
|
2qao |
B3110 |
Pout·4Bout |
|
|
|
B3708, B3709, B3710, B3711, B3712, B3713 |
|
U:B1267 |
|
A:B2009, U:B2011, G:B2012, G:B2010 |
|
2qba |
B3103 |
Pout·4Bout |
|
|
|
B3694, B3695, B3696, B3697, B3698, B3699 |
|
U:B1267 |
|
G:B2010, U:B2011, G:B2012, A:B2009 |
|
2qbc |
B3110 |
Pout·4Bout |
|
|
|
B3710, B3711, B3712, B3713, B3714, B3715 |
|
U:B1267 |
|
U:B2011, G:B2012, G:B2010, A:B2009 |
|
2qbe |
B3103 |
Pout·4Bout |
|
|
|
B3695, B3696, B3697, B3698, B3699, B3700 |
|
U:B1267 |
|
G:B2010, U:B2011, G:B2012, A:B2009 |
|
2qbg |
B2930 |
Pout·4Bout |
|
|
|
B3142, B3143, B3144, B3145, B3146, B3147 |
|
A:B2453 |
|
A:B2453, G:B2454, A:B2497, G:B2455 |
|
2qbg |
B2923 |
Pout·4Bout |
|
|
|
B3107, B3108, B3109, B3110, B3111, B3112 |
|
U:B1267 |
|
U:B2011, G:B2012, A:B2009, G:B2010 |
|
2qbi |
B2922 |
Pout·4Bout |
|
|
|
B3096, B3097, B3098, B3099, B3100, B3101 |
|
U:B1267 |
|
G:B2010, U:B2011, G:B2012, A:B2009 |
|
2qbj |
A1562 |
Pout·4Bout |
|
|
|
A1693, A1694, A1695, A1696, A1697, A1698 |
|
U:A644 |
|
G:A592, U:A593, G:A646, G:A645 |
|
2qbk |
B2923 |
Pout·4Bout |
|
|
|
B3106, B3107, B3108, B3109, B3110, B3111 |
|
U:B1267 |
|
A:B2009, U:B2011, G:B2012, G:B2010 |
|
2qox |
B2923 |
Pout·4Bout |
|
|
|
B3104, B3105, B3106, B3107, B3108, B3109 |
|
U:B1267 |
|
U:B2011, G:B2012, G:B2010, A:B2009 |
|