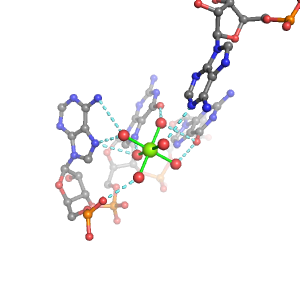
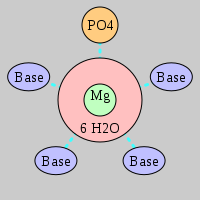
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2qoy |
A1548 |
Pout·4Bout |
|
|
|
A1924, A1925, A1926, A1927, A1928, A1929 |
|
U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
2qp1 |
B2939 |
Pout·4Bout |
|
|
|
B3691, B3692, B3693, B3694, B3695, B3696 |
|
A:B447 |
|
G:B473, U:B29, C:B31, G:B474 |
|
2z4k |
A1562 |
Pout·4Bout |
|
|
|
A1689, A1690, A1691, A1692, A1693 |
|
A:A1201 |
|
G:A1050, G:A1207, C:A1208, U:A1205 |
|
2z4k |
A1549 |
Pout·4Bout |
|
|
|
A1629, A1630, A1631, A1632, A1633, A1634 |
|
U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
2z4m |
A1563 |
Pout·4Bout |
|
|
|
A1692, A1693, A1694, A1695, A1696, A1697 |
|
U:A644 |
|
G:A592, U:A593, G:A646, G:A645 |
|
2z4n |
B2924 |
Pout·4Bout |
|
|
|
B3106, B3107, B3108, B3109, B3110, B3111 |
|
U:B1267 |
|
A:B2009, U:B2011, G:B2012, G:B2010 |
|
3df4 |
B3110 |
Pout·4Bout |
|
|
|
B4111, B4112, B4113, B4114, B4115, B4116 |
|
U:B1267 |
|
A:B2009, U:B2011, G:B2012, G:B2010 |
|
3i1m |
A1549 |
Pout·4Bout |
|
|
|
A1653, A1654, N121, N123, N124, N125 |
|
A:A1360 |
|
A:A1360, G:A1361, C:A979, C:A980 |
|
3i1p |
A2943 |
Pout·4Bout |
|
|
|
A3223, A3224, A3225, A3226, A3227, A3228 |
|
U:A1267 |
|
A:A2009, G:A2010, U:A2011, G:A2012 |
|
3i1p |
A2919 |
Pout·4Bout |
|
|
|
A3118, A3119, A3120, A3121, A3122, A3123 |
|
C:A1257 |
|
U:A580, C:A581, G:A1259, U:A1258 |
|
3i1q |
A1543 |
Pout·4Bout |
|
|
|
A1618, A1619, A1620, A1621, A1622, A1623 |
|
G:A645 |
|
G:A646, G:A592, U:A593, G:A645 |
|
3i1s |
A1558 |
Pout·4Bout |
|
|
|
A1708, A1709, A1710, A1711, A1712 |
|
A:A1349 |
|
A:A1350, G:A1371, U:A1372, G:A1370 |
|
3i1t |
A2925 |
Pout·4Bout |
|
|
|
A3148, A3149, A3150, A3151, A3152, A3153 |
|
U:A755 |
|
U:A755, G:A757, A:A739, A:A756 |
|
3i20 |
A2941 |
Pout·4Bout |
|
|
|
A3215, A3216, A3217, A3218, A3219, A3220 |
|
U:A1267 |
|
G:A2010, U:A2011, G:A2012, A:A2009 |
|
3i21 |
A1540 |
Pout·4Bout |
|
|
|
A1616, A1617, A1618, A1619, A1620, A1621 |
|
G:A645 |
|
C:A647, G:A645, G:A592, U:A593 |
|
3i22 |
A2979 |
Pout·4Bout |
|
|
|
A3399, A3400, A3401, A3402, A3403, A3404 |
|
G:A2454 |
|
C:A2496, G:A2454, G:A2455, A:A2453 |
|
3ivk |
C132 |
Pout·4Bout |
|
|
|
C177, C178, C179, C180, C181, C182 |
|
A:C-4 |
|
A:C-4, A:C14, G:C-3, U:C-2 |
|
3oar |
A1543 |
Pout·4Bout |
|
|
|
A1616, A1617, A1618, A1619, A1620, A1621 |
|
G:A645 |
|
G:A646, G:A645, G:A592, U:A593 |
|
3oas |
A2942 |
Pout·4Bout |
|
|
|
A3223, A3224, A3225, A3226, A3227, A3228 |
|
U:A1267 |
|
A:A2009, G:A2010, U:A2011, G:A2012 |
|
3oat |
A2984 |
Pout·4Bout |
|
|
|
A3409, A3410, A3411, A3412, A3413, A3414 |
|
A:A933 |
|
G:A841, U:A842, G:A843, A:A936 |
|
3ofa |
A1537 |
Pout·4Bout |
|
|
|
A1592, A1593, A1594, A1595, A1596, A1597 |
|
A:A415 |
|
G:A417, C:A418, G:A416, U:A426 |
|
3ofb |
A1543 |
Pout·4Bout |
|
|
|
A1617, A1618, A1619, A1620, A1621, A1622 |
|
G:A645 |
|
G:A646, G:A645, G:A592, U:A593 |
|
3ofc |
A2939 |
Pout·4Bout |
|
|
|
A3208, A3209, A3210, A3211, A3212, A3213 |
|
U:A1267 |
|
G:A2010, U:A2011, G:A2012, A:A2009 |
|
3ofd |
A2942 |
Pout·4Bout |
|
|
|
A3222, A3223, A3224, A3225, A3226, A3227 |
|
U:A1267 |
|
G:A2010, U:A2011, G:A2012, A:A2009 |
|
3ofo |
A1537 |
Pout·4Bout |
|
|
|
A1591, A1592, A1593, A1594, A1595, A1596 |
|
A:A415 |
|
G:A417, C:A418, G:A416, U:A426 |
|