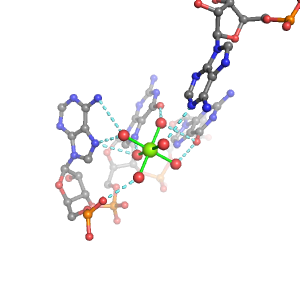
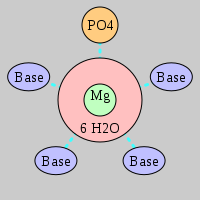
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3ofp |
A1537 |
Pout·4Bout |
|
|
|
A1589, A1590, A1591, A1592, A1593, A1594 |
|
A:A415 |
|
G:A417, G:A425, G:A416, U:A426 |
|
3ofr |
A2986 |
Pout·4Bout |
|
|
|
A3419, A3420, A3421, A3422, A3423, A3424 |
|
A:A933 |
|
G:A843, G:A841, U:A842, A:A936 |
|
3ofy |
A1543 |
Pout·4Bout |
|
|
|
A1617, A1618, A1619, A1620, A1621, A1622 |
|
G:A645 |
|
G:A646, G:A645, G:A592, U:A593 |
|
3og0 |
A2924 |
Pout·4Bout |
|
|
|
A3143, A3144, A3145, A3146, A3147, A3148 |
|
U:A755 |
|
U:A755, G:A757, A:A739, A:A756 |
|
3og0 |
A2978 |
Pout·4Bout |
|
|
|
A3396, A3397, A3398, A3399, A3400, A3401 |
|
G:A2454 |
|
G:A2454, G:A2455, C:A2496, A:A2453 |
|
3og0 |
A2941 |
Pout·4Bout |
|
|
|
A3222, A3223, A3224, A3225, A3226, A3227 |
|
U:A1267 |
|
A:A2009, G:A2010, U:A2011, G:A2012 |
|
3ora |
A1561 |
Pout·4Bout |
|
|
|
A1712, A1713, A1714, A1715, A1716 |
|
A:A1349 |
|
U:A1351, G:A1371, U:A1372, G:A1370 |
|
3r8t |
A3034 |
Pout·4Bout |
|
|
|
A3659, A3660, A3661, A3662, A3663, A3664 |
|
C:A1790 |
|
G:A1792, U:A1827, G:A1826, C:A1793 |
|
3v23 |
A3602 |
Pout·4Bout |
|
|
|
A5006, A5007, A5008, A5009, A5010, A5011 |
|
A:A103 |
|
G:A80, G:A81, G:A79, G:A82 |
|
3v26 |
A1711 |
Pout·4Bout |
|
|
|
A1947, A1948, A1949, A1950, A1951, A1952 |
|
C:A398 |
|
C:A36, U:A37, G:A35, G:A548 |
|
3v27 |
A3415 |
Pout·4Bout |
|
|
|
A4284, A4285, A4286, A4287, A4288 |
|
G:A186 |
|
G:A186, G:A187, G:A189, G:A188 |
|
3v29 |
A3572 |
Pout·4Bout |
|
|
|
A4661, A4662, A4663, A4664, A4665, A4666 |
|
U:A1267 |
|
G:A2010, G:A2009, U:A2011, G:A2012 |
|
3v2d |
A3659 |
Pout·4Bout |
|
|
|
A5537, A5538, A5539, A5540, A5541 |
|
A:A1701 |
|
G:A1703, G:A1702, G:A1699, G:A1763 |
|
3v2d |
A3684 |
Pout·4Bout |
|
|
|
A5625, A5626, A5627, A5628, A5629 |
|
G:A686 |
|
G:A690, G:A771, U:A688, A:A689 |
|
4a1a |
0204 |
Pout·4Bout |
|
|
|
06675, 06676, 06677, 06678, 06679, 06680 |
|
A:374 |
|
U:376, A:3101, G:3102, A:3100 |
|
4a1c |
0204 |
Pout·4Bout |
|
|
|
06291, 06292, 06293, 06294, 06295, 06296 |
|
A:374 |
|
U:376, A:3101, G:3102, A:3100 |
|
4bpe |
A5009 |
Pout·4Bout |
|
|
|
A2106, A2107, A2119, A2120, A2122, A2253 |
|
C:A577 |
|
G:A550, G:A580, C:A578, A:A579 |
|
4bpn |
A5009 |
Pout·4Bout |
|
|
|
A6189, A6197, A6200, A6207, A6214, A6221 |
|
C:A577 |
|
A:A579, G:A580, G:A550, C:A552 |
|
4dh9 |
A1870 |
Pout·4Bout |
|
|
|
A2091, A2092, A2093, A2094 |
|
A:A583 |
|
G:A584, G:A585, C:A756, U:A757 |
|
4dha |
A3710 |
Pout·4Bout |
|
|
|
A4766, A4767, A4768, A4769, A4770, A4771 |
|
A:A2360 |
|
U:A2357, A:A2361, C:A2356, G:A2362 |
|
4dha |
A3604 |
Pout·4Bout |
|
|
|
A4380, A4381, A4382, A4383, A4384 |
|
G:A686 |
|
G:A771, U:A688, A:A689, G:A690 |
|
4dhc |
A3581 |
Pout·4Bout |
|
|
|
A4392, A4393, A4394, A4395, A4396 |
|
G:A495 |
|
A:A482, G:A496, G:A494, G:A495 |
|
4dr2 |
A1751 |
Pout·4Bout |
|
|
|
A2388, A2389, A2390, A2391 |
|
A:A1360 |
|
C:A980, C:A979, A:A1360, G:A1361 |
|
4dr2 |
A1665 |
Pout·4Bout |
|
|
|
A2135, A2136, A2137, A2138, A2139 |
|
A:A583 |
|
G:A584, G:A585, C:A756, U:A757 |
|
4dr4 |
A1942 |
Pout·4Bout |
|
|
|
A2404, A2405, A2406, A2407, A2408, A2409 |
|
A:A642 |
|
G:A595, G:A594, G:A593, G:A592 |
|