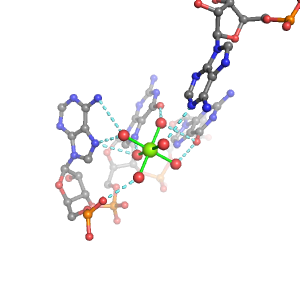
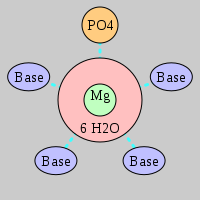
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4pea |
A1620 |
Pout·4Bout |
|
|
|
A1788, A1789, A1790, A1791, A1792, A1793 |
|
G:A645 |
|
G:A645, G:A646, G:A592, U:A593 |
|
4pea |
A1605 |
Pout·4Bout |
|
|
|
A1718, A1719, A1720, A1721, A1722, A1723 |
|
G:A266 |
|
G:A257, G:A258, G:A259, C:A267 |
|
4peb |
A3070 |
Pout·4Bout |
|
|
|
A3527, A3528, A3529, A3530, A3531, A3532 |
|
A:A2453 |
|
A:A2453, G:A2454, A:A2497, G:A2455 |
|
4pec |
A3102 |
Pout·4Bout |
|
|
|
A3669, A3670, A3671, A3672, A3673, A3674 |
|
C:A2616 |
|
G:A2049, C:A2047, G:A2048, G:A2618 |
|
4qcn |
A3653 |
Pout·4Bout |
|
|
|
A5082, A5083, A5084, A5085, A5086, A5087 |
|
A:A1748 |
|
G:A1749, G:A1746, G:A1750, G:A1794 |
|
4qct |
A3383 |
Pout·4Bout |
|
|
|
A4364, A4365, A4366, A4367, A4368 |
|
G:A2845 |
|
G:A2848, U:A2847, G:A2845, G:A2846 |
|
4qcx |
A3379 |
Pout·4Bout |
|
|
|
A4287, A4288, A4289, A4290, A4291 |
|
A:A2453 |
|
A:A2453, A:A2497, G:A2454, G:A2455 |
|
4qcx |
A3384 |
Pout·4Bout |
|
|
|
A4300, A4301, A4302, A4303, A4304 |
|
G:A2845 |
|
G:A2848, G:A2846, U:A2847, G:A2845 |
|
4qcz |
A3513 |
Pout·4Bout |
|
|
|
A4662, A4663, A4664, A4665, A4666, A4667 |
|
A:A101 |
|
G:A79, G:A80, G:A78, G:A81 |
|
4qcz |
A3651 |
Pout·4Bout |
|
|
|
A4986, A4987, A4988, A4989, A4990, A4991 |
|
A:A1748 |
|
G:A1794, G:A1746, G:A1749, G:A1750 |
|
4tom |
A3083 |
Pout·4Bout |
|
|
|
A3582, A3583, A3584, A3585, A3586, A3587 |
|
A:A933 |
|
G:A841, U:A842, G:A843, A:A936 |
|
4ton |
A1612 |
Pout·4Bout |
|
|
|
A1751, A1752, A1753, A1754, A1755, A1756 |
|
A:A415 |
|
G:A417, G:A425, U:A426, G:A416 |
|
4too |
A3094 |
Pout·4Bout |
|
|
|
A3625, A3626, A3627, A3628 |
|
A:A1439 |
|
A:A1552, U:A1440, G:A1441, A:A1551 |
|
4tou |
A1604 |
Pout·4Bout |
|
|
|
A1716, A1717, A1718, A1719, A1720, A1721 |
|
A:A415 |
|
G:A416, G:A417, C:A418, G:A425 |
|
4tou |
A1610 |
Pout·4Bout |
|
|
|
A1744, A1745, A1746, A1747, A1748, A1749 |
|
U:A644 |
|
G:A646, U:A593, U:A594, G:A645 |
|
4tow |
A1630 |
Pout·4Bout |
|
|
|
A1839, A1840, A1841, A1842, N201, N202 |
|
A:A1360 |
|
A:A1360, G:A1361, C:A979, C:A980 |
|
4tow |
A1612 |
Pout·4Bout |
|
|
|
A1750, A1751, A1752, A1753, A1754, A1755 |
|
A:A415 |
|
G:A417, G:A425, U:A426, G:A416 |
|
4tox |
A3019 |
Pout·4Bout |
|
|
|
A3286, A3287, A3288, A3289, A3290, A3291 |
|
A:A616 |
|
G:A617, G:A619, G:A618, C:A610 |
|
4tp0 |
A1610 |
Pout·4Bout |
|
|
|
A1744, A1745, A1746, A1747, A1748, A1749 |
|
G:A645 |
|
G:A646, U:A593, U:A594, G:A645 |
|
4tp2 |
A1605 |
Pout·4Bout |
|
|
|
A1718, A1719, A1720, A1721, A1722, A1723 |
|
G:A266 |
|
G:A257, G:A258, G:A259, C:A267 |
|
4tp3 |
A3103 |
Pout·4Bout |
|
|
|
A3669, A3670, A3671, A3672, A3673, A3674 |
|
C:A2616 |
|
G:A2049, G:A2618, C:A2047, G:A2048 |
|
4tp4 |
A1622 |
Pout·4Bout |
|
|
|
A1804, A1805, A1806, A1807, A1808, A1809 |
|
G:A266 |
|
G:A257, G:A259, G:A258, U:A268 |
|
4tp5 |
A3071 |
Pout·4Bout |
|
|
|
A3525, A3526, A3527, A3528, A3529, A3530 |
|
C:A2452 |
|
A:A2453, G:A2454, A:A2497, G:A2455 |
|
4tp6 |
A1605 |
Pout·4Bout |
|
|
|
A1718, A1719, A1720, A1721, A1722, A1723 |
|
G:A266 |
|
G:A257, G:A258, G:A259, C:A267 |
|
4tp6 |
A1612 |
Pout·4Bout |
|
|
|
A1752, A1753, A1754, A1755, A1756, A1757 |
|
A:A415 |
|
G:A417, U:A426, G:A425, G:A416 |
|