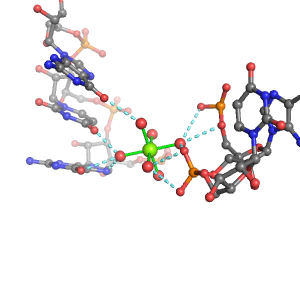
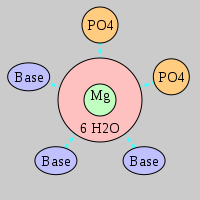
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1jj2 |
08071 |
2Pout·3Bout |
|
|
|
03639, 05598, 06971, 07112, 07385, 07386 |
|
A:01815, C:01816 |
|
U:01813, C:01818, U:01817 |
|
1k73 |
A8071 |
2Pout·3Bout |
|
|
|
A4108, A6060, A7436, A7573, A7849, A7850 |
|
A:A1815, C:A1816 |
|
U:A1813, U:A1817, C:A1818 |
|
1k8a |
A8071 |
2Pout·3Bout |
|
|
|
A3699, A5624, A6978, A7112, A7387, A7388 |
|
A:A1815, C:A1816 |
|
U:A1813, U:A1817, C:A1818 |
|
1k9m |
A8071 |
2Pout·3Bout |
|
|
|
A4107, A6058, A7434, A7571, A7848, A7849 |
|
A:A1815, C:A1816 |
|
U:A1813, U:A1817, C:A1818 |
|
1kc8 |
A8071 |
2Pout·3Bout |
|
|
|
A4116, A6051, A7425, A7561, A7837, A7838 |
|
A:A1815, C:A1816 |
|
U:A1813, U:A1817, C:A1818 |
|
1kd1 |
A8071 |
2Pout·3Bout |
|
|
|
A4105, A6048, A7421, A7555, A7835, A7836 |
|
A:A1815, C:A1816 |
|
U:A1813, C:A1818, U:A1817 |
|
1m1k |
A8071 |
2Pout·3Bout |
|
|
|
A3708, A5657, A7015, A7150, A7426, A7427 |
|
A:A1815, C:A1816 |
|
U:A1813, U:A1817, C:A1818 |
|
1m90 |
A8071 |
2Pout·3Bout |
|
|
|
A3642, A5592, A6955, A7096, A7368, A7369 |
|
A:A1815, C:A1816 |
|
U:A1813, C:A1818, U:A1817 |
|
1n78 |
D701 |
2Pout·3Bout |
|
|
|
D2323, D2324, D2325, D2326, D2327, D2328 |
|
A:D546, G:D522 |
|
A:D546, A:D545, C:D509 |
|
1n78 |
C700 |
2Pout·3Bout |
|
|
|
C2317, C2318, C2319, C2320, C2321, C2322 |
|
A:C546, G:C522 |
|
C:C509, A:C545, A:C546 |
|
1nji |
A8071 |
2Pout·3Bout |
|
|
|
A4103, A6053, A7417, A7553, A7830, A7831 |
|
A:A1815, C:A1816 |
|
U:A1813, U:A1817, C:A1818 |
|
1nuj |
B419 |
2Pout·3Bout |
|
|
|
A426, A427, B425, B426, B427, B428 |
|
G:A26, A:A25 |
|
U:B41, G:B42, G:B43 |
|
1nuj |
H422 |
2Pout·3Bout |
|
|
|
G235, G238, H234, H236, H237, H239 |
|
G:G26, A:G25 |
|
U:H41, G:H42, G:H43 |
|
1nuv |
B412 |
2Pout·3Bout |
|
|
|
A421, A422, A423, B428, B429, B430 |
|
G:A26, A:A25 |
|
U:B41, G:B42, G:B43 |
|
1nuv |
H415 |
2Pout·3Bout |
|
|
|
G228, G229, G231, H227, H230, H232 |
|
G:G26, A:G25 |
|
U:H41, G:H42, G:H43 |
|
1q7y |
A8071 |
2Pout·3Bout |
|
|
|
A3620, A5548, A6906, A7040, A7310, A7311 |
|
A:A1815, C:A1816 |
|
U:A1813, C:A1818, U:A1817 |
|
1q81 |
A8071 |
2Pout·3Bout |
|
|
|
A3622, A5569, A6910, A7049, A7319, A7320 |
|
A:A1815, C:A1816 |
|
U:A1813, C:A1818, U:A1817 |
|
1q82 |
A8071 |
2Pout·3Bout |
|
|
|
A3624, A5539, A6901, A7035, A7310, A7311 |
|
A:A1815, C:A1816 |
|
U:A1817, C:A1818, U:A1813 |
|
1q86 |
A8071 |
2Pout·3Bout |
|
|
|
A3641, A5571, A6936, A7066, A7340, A7341 |
|
A:A1815, C:A1816 |
|
U:A1813, U:A1817, C:A1818 |
|
1qvf |
08071 |
2Pout·3Bout |
|
|
|
03623, 05541, 06891, 07021, 07286, 07287 |
|
A:01815, C:01816 |
|
U:01813, C:01818, U:01817 |
|
1s72 |
08071 |
2Pout·3Bout |
|
|
|
03636, 05579, 06933, 07068, 07340, 07341 |
|
A:01815, C:01816 |
|
U:01813, C:01818, U:01817 |
|
1vq4 |
08071 |
2Pout·3Bout |
|
|
|
04424, 06318, 07618, 07743, 08065, 08066 |
|
A:01815, C:01816 |
|
U:01813, C:01818, U:01817 |
|
1vs5 |
A1546 |
2Pout·3Bout |
|
|
|
A1615, A1616, A1617, A1618, A1619, A1620 |
|
G:A803, U:A804 |
|
U:A772, G:A773, G:A771 |
|
1vs6 |
B2923 |
2Pout·3Bout |
|
|
|
B3104, B3105, B3106, B3107, B3108, B3109 |
|
U:B29, A:B447 |
|
G:B474, U:B29, G:B30 |
|
1vs7 |
A1547 |
2Pout·3Bout |
|
|
|
A1618, A1619, A1620, A1621, A1622, A1623 |
|
G:A803, U:A804 |
|
U:A772, G:A773, G:A771 |
|