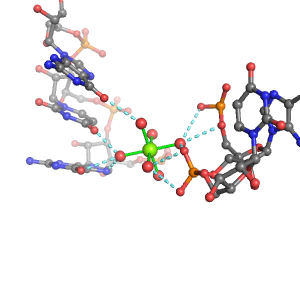
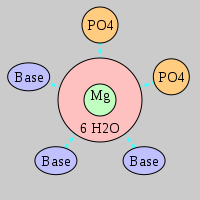
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1vs8 |
B2927 |
2Pout·3Bout |
|
|
|
B3124, B3125, B3126, B3127, B3128, B3129 |
|
A:B616, G:B617 |
|
G:B617, G:B618, G:B619 |
|
1yi2 |
08071 |
2Pout·3Bout |
|
|
|
04108, 06035, 07392, 07524, 07795, 07796 |
|
A:01815, C:01816 |
|
U:01813, C:01818, U:01817 |
|
1yij |
08071 |
2Pout·3Bout |
|
|
|
04127, 06056, 07408, 07537, 07808, 07809 |
|
A:01815, C:01816 |
|
U:01813, C:01818, U:01817 |
|
1yjn |
08071 |
2Pout·3Bout |
|
|
|
04128, 06069, 07418, 07549, 07821, 07822 |
|
A:01815, C:01816 |
|
U:01813, C:01818, U:01817 |
|
1yls |
B245 |
2Pout·3Bout |
|
|
|
B373, B374, B375, B376, B377, B378 |
|
A:B206, G:B205 |
|
A:B206, G:B205, G:B207 |
|
1yls |
D342 |
2Pout·3Bout |
|
|
|
D399, D400, D401, D402, D403, D404 |
|
C:D215, G:D216 |
|
G:D217, G:D227, G:D216 |
|
2avy |
A1545 |
2Pout·3Bout |
|
|
|
A1613, A1614, A1615, A1616, A1617, A1618 |
|
G:A803, U:A804 |
|
U:A772, G:A773, G:A771 |
|
2aw4 |
B2957 |
2Pout·3Bout |
|
|
|
B3262, B3263, B3264, B3265, B3266, B3267 |
|
A:B616, G:B617 |
|
G:B617, G:B618, G:B619 |
|
2aw4 |
B2923 |
2Pout·3Bout |
|
|
|
B3101, B3102, B3103, B3104, B3105, B3106 |
|
U:B29, A:B447 |
|
G:B474, U:B29, G:B30 |
|
2aw7 |
A1546 |
2Pout·3Bout |
|
|
|
A1617, A1618, A1619, A1620, A1621, A1622 |
|
G:A803, U:A804 |
|
U:A772, G:A773, G:A771 |
|
2awb |
B2927 |
2Pout·3Bout |
|
|
|
B3125, B3126, B3127, B3128, B3129, B3130 |
|
G:B617, A:B616 |
|
G:B617, G:B618, G:B619 |
|
2i2p |
X57 |
2Pout·3Bout |
|
|
|
A1877, A1878, X341, X342, X344, X346 |
|
G:X3, U:X2 |
|
C:A1399, G:A1401, G:X3 |
|
2i2p |
A1597 |
2Pout·3Bout |
|
|
|
A1866, A1867, A1868, A1869, A1870 |
|
C:A110, G:A111 |
|
G:A112, G:A111, G:A113 |
|
2i2t |
B2923 |
2Pout·3Bout |
|
|
|
B3111, B3112, B3113, B3114, B3115, B3116 |
|
U:B29, A:B447 |
|
G:B474, U:B29, G:B30 |
|
2i2t |
B3001 |
2Pout·3Bout |
|
|
|
B3461, B3462, B3463, B3464, B3465, B3466 |
|
A:B1000, A:B1001 |
|
G:B1003, G:B1002, C:B1153 |
|
2i2t |
B2983 |
2Pout·3Bout |
|
|
|
B3394, B3395, B3396, B3397, B3398, B3399 |
|
A:B2453, C:B2452 |
|
A:B2453, A:B2497, G:B2454 |
|
2nz4 |
Q9007 |
2Pout·3Bout |
|
|
|
F33, F35, Q9013, Q9014, Q9015, Q9016 |
|
C:Q52, G:Q53 |
|
G:Q54, A:Q28, G:Q53 |
|
2otl |
08071 |
2Pout·3Bout |
|
|
|
04130, 06070, 07417, 07550, 07818, 07819 |
|
A:01815, C:01816 |
|
U:01813, U:01817, C:01818 |
|
2qam |
B3309 |
2Pout·3Bout |
|
|
|
B3863, B3864, B3865, B3866, B3867, B3868 |
|
A:B616, G:B617 |
|
G:B617, G:B618, G:B619 |
|
2qb9 |
A2014 |
2Pout·3Bout |
|
|
|
A2370, A2371, A2372, A2373, A2374, A2375 |
|
G:A803, U:A804 |
|
U:A772, G:A773, G:A771 |
|
2qb9 |
A2044 |
2Pout·3Bout |
|
|
|
A2395, A2396, A2397, A2398, A2399, A2400 |
|
A:A315, A:A329 |
|
G:A319, A:A320, G:A318 |
|
2qbb |
A2016 |
2Pout·3Bout |
|
|
|
A2382, A2383, A2384, A2385, A2386, A2387 |
|
G:A803, U:A804 |
|
U:A772, G:A773, G:A771 |
|
2qbc |
B3133 |
2Pout·3Bout |
|
|
|
B3729, B3730, B3731, B3732, B3733, B3734 |
|
A:B616, G:B617 |
|
G:B618, G:B619, G:B617 |
|
2qbe |
B3309 |
2Pout·3Bout |
|
|
|
B3863, B3864, B3865, B3866, B3867, B3868 |
|
A:B616, G:B617 |
|
G:B617, G:B618, G:B619 |
|
2qbg |
B2927 |
2Pout·3Bout |
|
|
|
B3126, B3127, B3128, B3129, B3130, B3131 |
|
A:B616, G:B617 |
|
G:B617, G:B618, G:B619 |
|