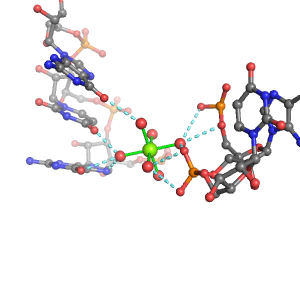
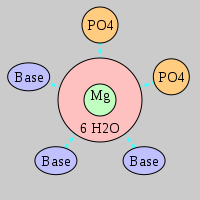
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3i1n |
A3037 |
2Pout·3Bout |
|
|
|
A3638, A3639, A3640, A3641, 4779, 4784 |
|
A:A2740, A:A2741 |
|
G:A2742, G:A2763, U:A2743 |
|
3i1n |
A2921 |
2Pout·3Bout |
|
|
|
A3129, A3130, A3131, A3132, A3133, A3134 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3i1p |
A2921 |
2Pout·3Bout |
|
|
|
A3130, A3131, A3132, A3133, A3134, A3135 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3i1p |
A2978 |
2Pout·3Bout |
|
|
|
A3386, A3387, A3388, A3389, A3390, A3391 |
|
U:A2387, A:A2386 |
|
C:A2326, G:A2389, U:A2387 |
|
3i1q |
A1535 |
2Pout·3Bout |
|
|
|
A1581, A1582, A1583, A1584, A1585, A1586 |
|
A:A329, A:A315 |
|
A:A320, G:A318, G:A319 |
|
3i1r |
A2921 |
2Pout·3Bout |
|
|
|
A3128, A3129, A3130, A3131, A3132, E235 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3i1s |
A4 |
2Pout·3Bout |
|
|
|
A1588, A1589, A1590, A1591, A1592, A1593 |
|
A:A415, G:A416 |
|
G:A417, G:A425, G:A416 |
|
3i1s |
A2 |
2Pout·3Bout |
|
|
|
A1577, A1578, A1579, A1580, A1581, A1582 |
|
A:A315, A:A329 |
|
A:A320, G:A318, G:A319 |
|
3i1t |
A3003 |
2Pout·3Bout |
|
|
|
A3507, A3508, A3509, A3510, A3511 |
|
A:A2705, C:A2704 |
|
A:A2705, U:A2701, A:A2706 |
|
3i1t |
A2921 |
2Pout·3Bout |
|
|
|
A3129, A3130, A3131, A3132, A3133, A3134 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3i1z |
A1537 |
2Pout·3Bout |
|
|
|
A1593, A1594, A1595, A1596, A1597, A1598 |
|
A:A415, G:A416 |
|
G:A417, C:A418, G:A416 |
|
3i20 |
A2921 |
2Pout·3Bout |
|
|
|
A3129, A3130, A3131, A3132, A3133, A3134 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3i20 |
A2999 |
2Pout·3Bout |
|
|
|
A3488, A3489, A3490, A3491, A3492, A3493 |
|
C:A1617, G:A1619 |
|
G:A1619, G:A1620, G:A1613 |
|
3i21 |
A4 |
2Pout·3Bout |
|
|
|
A1588, A1589, A1590, A1591, A1592, A1593 |
|
A:A415, G:A416 |
|
G:A417, C:A418, G:A416 |
|
3oar |
A1544 |
2Pout·3Bout |
|
|
|
A1622, A1623, A1624, A1625, A1626, A1627 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
3oar |
A1537 |
2Pout·3Bout |
|
|
|
A1588, A1589, A1590, A1591, A1592, A1593 |
|
A:A415, G:A416 |
|
G:A417, G:A425, G:A416 |
|
3oas |
A3002 |
2Pout·3Bout |
|
|
|
A3506, A3507, A3508, A3509, A3510 |
|
A:A2705, C:A2704 |
|
A:A2705, A:A2706, U:A2707 |
|
3oat |
A2920 |
2Pout·3Bout |
|
|
|
A3120, A3121, A3122, A3123, A3124, A3125 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3ofb |
A1544 |
2Pout·3Bout |
|
|
|
A1623, A1624, A1625, A1626, A1627, A1628 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
3ofb |
A1561 |
2Pout·3Bout |
|
|
|
A1707, A1708, A1709, A1710, I169 |
|
U:A1348, A:A1349 |
|
G:A1371, U:A1372, G:A1370 |
|
3ofb |
A1537 |
2Pout·3Bout |
|
|
|
A1589, A1590, A1591, A1592, A1593, A1594 |
|
A:A415, G:A416 |
|
G:A417, G:A416, U:A426 |
|
3ofc |
A3035 |
2Pout·3Bout |
|
|
|
A3632, A3633, A3634, A3635, 4769, 4774 |
|
A:A2740, A:A2741 |
|
G:A2742, G:A2763, U:A2743 |
|
3ofc |
A3037 |
2Pout·3Bout |
|
|
|
A3639, A3640, A3641, A3642, A3643, A3644 |
|
A:A1650, G:A2004 |
|
G:A1651, G:A1649, A:A1650 |
|
3ofd |
A3002 |
2Pout·3Bout |
|
|
|
A3505, A3506, A3507, A3508, A3509 |
|
A:A2705, C:A2704 |
|
A:A2705, A:A2706, U:A2707 |
|
3ofp |
A1544 |
2Pout·3Bout |
|
|
|
A1623, A1624, A1625, A1626, A1627, A1628 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|