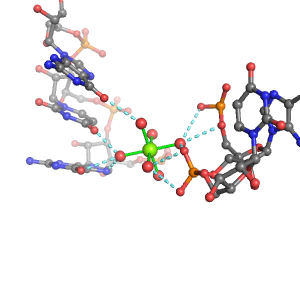
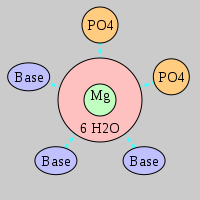
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3ofr |
A2921 |
2Pout·3Bout |
|
|
|
A3123, A3124, A3125, A3126, A3127, E202 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3ofx |
A1544 |
2Pout·3Bout |
|
|
|
A1626, A1627, A1628, A1629, A1630, A1631 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
3ofy |
A1537 |
2Pout·3Bout |
|
|
|
A1589, A1590, A1591, A1592, A1593, A1594 |
|
A:A415, G:A416 |
|
G:A417, G:A416, U:A426 |
|
3ofy |
A1549 |
2Pout·3Bout |
|
|
|
A1648, A1649, A1650, A1651, N101, N102 |
|
A:A1360, C:A1359 |
|
C:A980, G:A1361, A:A1360 |
|
3ofy |
A1544 |
2Pout·3Bout |
|
|
|
A1623, A1624, A1625, A1626, A1627, A1628 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
3ofz |
A2920 |
2Pout·3Bout |
|
|
|
A3119, A3120, A3121, A3122, A3123, A3124 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3ora |
A1537 |
2Pout·3Bout |
|
|
|
A1593, A1594, A1595, A1596, A1597, A1598 |
|
A:A415, G:A416 |
|
G:A417, G:A425, G:A416 |
|
3ora |
A1544 |
2Pout·3Bout |
|
|
|
A1627, A1628, A1629, A1630, A1631, A1632 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
3ora |
A1535 |
2Pout·3Bout |
|
|
|
A1582, A1583, A1584, A1585, A1586, A1587 |
|
A:A315, A:A329 |
|
G:A319, A:A320, G:A318 |
|
3orb |
A2921 |
2Pout·3Bout |
|
|
|
A3124, A3125, A3126, A3127, A3128, A3129 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
3q3z |
A83 |
2Pout·3Bout |
|
|
|
A103, A105, A106, A113, A115, A116 |
|
G:A73, C:A72 |
|
G:A12, A:A74, G:A73 |
|
3r8s |
A2996 |
2Pout·3Bout |
|
|
|
A3528, A3529, A3530, A3531, A3532, A3533 |
|
A:A1572, G:A1422 |
|
G:A1423, G:A1424, G:A1425 |
|
3r8s |
A2921 |
2Pout·3Bout |
|
|
|
A3186, A3187, A3188, A3189, A3190, A3191 |
|
G:A617, A:A616 |
|
G:A617, G:A618, G:A619 |
|
3r8t |
A2995 |
2Pout·3Bout |
|
|
|
A3499, A3500, A3501, A3502, A3503, A3504 |
|
A:A1572, G:A1422 |
|
G:A1423, G:A1425, G:A1424 |
|
3r8t |
A3036 |
2Pout·3Bout |
|
|
|
A3668, A3669, A3670, A3671, A3672, A3673 |
|
A:A2740, A:A2741 |
|
G:A2763, G:A2742, U:A2743 |
|
3v25 |
A3425 |
2Pout·3Bout |
|
|
|
A4179, A4180, A4181, A4182 |
|
C:A2452, A:A2453 |
|
A:A2453, G:A2454, A:A2497 |
|
3v27 |
A3321 |
2Pout·3Bout |
|
|
|
A3904, A3905, A3906, A3907, A3908, A3909 |
|
A:A299, C:A336 |
|
G:A338, A:A299, A:A300 |
|
3v27 |
A3409 |
2Pout·3Bout |
|
|
|
A4260, A4261, A4262, A4263, A4264 |
|
U:A1397, C:A1398 |
|
C:A1386, G:A1401, G:A1400 |
|
3v27 |
A3423 |
2Pout·3Bout |
|
|
|
A4315, A4316, A4317, A4318, A4319, A4320 |
|
A:A1634, G:A1627 |
|
G:A1628, U:A1629, G:A1630 |
|
3v29 |
A3433 |
2Pout·3Bout |
|
|
|
A4151, A4152, A4153, A4154, A4155, A4156 |
|
A:A2360, C:A2359 |
|
A:A2361, U:A2357, A:A2360 |
|
3v29 |
A3425 |
2Pout·3Bout |
|
|
|
A4121, A4122, A4123, A4124, A4125 |
|
A:A1572, G:A1421 |
|
G:A1423, G:A1424, G:A1425 |
|
3v29 |
A3574 |
2Pout·3Bout |
|
|
|
A4673, A4674, A4675, A4676, A4677, A4678 |
|
C:A1398, U:A1397 |
|
G:A1401, G:A1400, G:A1385 |
|
3v2c |
A1680 |
2Pout·3Bout |
|
|
|
A2036, A2037, A2038, A2039 |
|
U:A582, A:A583 |
|
G:A585, G:A584, U:A757 |
|
3v2d |
A3499 |
2Pout·3Bout |
|
|
|
A5123, A5124, A5125, A5126, A5127 |
|
A:A2005, G:A2004 |
|
G:A1650, G:A1651, G:A1649 |
|
3v2d |
A3554 |
2Pout·3Bout |
|
|
|
A5274, A5275, A5276, A5277, A5278, A5279 |
|
A:A2360, C:A2359 |
|
U:A2357, A:A2361, C:A2356 |
|