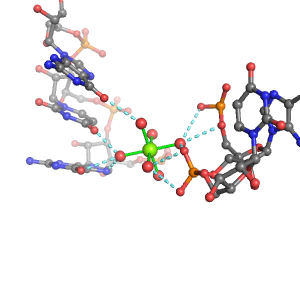
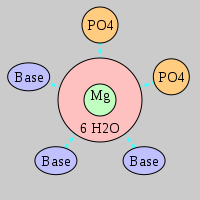
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4gas |
A1622 |
2Pout·3Bout |
|
|
|
A1802, A1803, A1804, A1805, A1806, A1807 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
4gau |
A3089 |
2Pout·3Bout |
|
|
|
A3626, A3627, A3628, A3629, A3630, A3631 |
|
G:A1422, A:A1572 |
|
G:A1423, G:A1425, G:A1424 |
|
4gau |
A3081 |
2Pout·3Bout |
|
|
|
A3588, A3589, A3590, A3591, A3592, A3593 |
|
G:A976, G:A977 |
|
G:A978, A:A979, G:A977 |
|
4gd2 |
A1608 |
2Pout·3Bout |
|
|
|
A1734, A1735, A1736, A1737, A1738, A1739 |
|
A:A315, A:A329 |
|
G:A319, A:A320, G:A318 |
|
4gd2 |
A1622 |
2Pout·3Bout |
|
|
|
A1803, A1804, A1805, A1806, A1807, A1808 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
4ji1 |
A1788 |
2Pout·3Bout |
|
|
|
A2420, A2421, A2422, A2423, A2424, A2425 |
|
G:A266, G:A265 |
|
G:A257, G:A258, G:A259 |
|
4ji5 |
A1700 |
2Pout·3Bout |
|
|
|
A1929, A1930, A1931, A1932, A1933, A1934 |
|
U:A804, G:A803 |
|
G:A771, U:A772, C:A770 |
|
4ji6 |
A1803 |
2Pout·3Bout |
|
|
|
A2445, A2446, A2447, A2448, A2449, A2450 |
|
U:A804, G:A803 |
|
G:A771, U:A772, C:A770 |
|
4ji7 |
A1850 |
2Pout·3Bout |
|
|
|
A2617, A2618, A2619, A2620, A2621, A2622 |
|
U:A804, G:A803 |
|
G:A771, U:A772, C:A770 |
|
4ji8 |
A1832 |
2Pout·3Bout |
|
|
|
A2514, A2515, A2516, A2517, A2518, A2519 |
|
U:A804, G:A803 |
|
G:A771, U:A772, C:A770 |
|
4nvy |
A3145 |
2Pout·3Bout |
|
|
|
A4011, A4012, A4013, A4014 |
|
U:A582, A:A583 |
|
G:A585, G:A584, U:A757 |
|
4pea |
A1606 |
2Pout·3Bout |
|
|
|
A1724, A1725, A1726, A1727, T101, T102 |
|
G:A258, G:A259 |
|
G:A258, G:A259, G:A260 |
|
4pea |
A1622 |
2Pout·3Bout |
|
|
|
A1799, A1800, A1801, A1802, A1803, A1804 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
4pec |
A3081 |
2Pout·3Bout |
|
|
|
A3568, A3569, A3570, A3571, A3572, 0101 |
|
C:A2055, A:A574 |
|
U:A2034, G:A2035, A:A574 |
|
4pec |
A3018 |
2Pout·3Bout |
|
|
|
A3285, A3286, A3287, A3288, A3289, E304 |
|
A:A608, A:A616 |
|
G:A617, G:A619, G:A618 |
|
4qcn |
A3308 |
2Pout·3Bout |
|
|
|
A4116, A4117, A4118, A4119, A4120, A4121 |
|
A:A323, C:A360 |
|
A:A324, A:A323, G:A362 |
|
4qcq |
A3170 |
2Pout·3Bout |
|
|
|
A4157, A4158, A4159, V102, V103 |
|
G:A1401, G:V18 |
|
G:A1401, C:A1399, G:V18 |
|
4qcr |
A3718 |
2Pout·3Bout |
|
|
|
A5193, A5194, A5195, A5196, A5197 |
|
G:A733, C:A734 |
|
U:A735, G:A818, G:A737 |
|
4qcr |
A3327 |
2Pout·3Bout |
|
|
|
A4130, A4131, A4132, A4133, A4134, A4135 |
|
A:A323, C:A360 |
|
A:A324, A:A323, G:A362 |
|
4qct |
A3268 |
2Pout·3Bout |
|
|
|
A4067, A4068, A4069, A4070, A4071, A4072 |
|
A:A299, C:A336 |
|
A:A300, A:A299, G:A338 |
|
4qcv |
A3296 |
2Pout·3Bout |
|
|
|
A4124, A4125, A4126, A4127, A4128, A4129 |
|
A:A323, C:A360 |
|
A:A324, A:A323, G:A362 |
|
4qcz |
A3309 |
2Pout·3Bout |
|
|
|
A4117, A4118, A4119, A4120, A4121, A4122 |
|
A:A323, C:A360 |
|
A:A324, A:A323, G:A362 |
|
4qd1 |
A3517 |
2Pout·3Bout |
|
|
|
A4527, A4528, A4529, A4530, A4531, A4532 |
|
A:A2435, A:A2434 |
|
U:A2431, U:A827, A:A2426 |
|
4qd1 |
A3296 |
2Pout·3Bout |
|
|
|
A4060, A4061, A4062, A4063, A4064, A4065 |
|
A:A299, C:A336 |
|
A:A300, G:A338, A:A299 |
|
4tol |
A1602 |
2Pout·3Bout |
|
|
|
A1706, A1707, A1708, A1709, A1710, A1711 |
|
A:A329, A:A315 |
|
A:A320, G:A318, G:A319 |
|