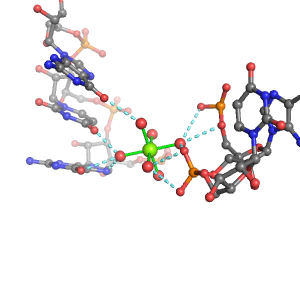
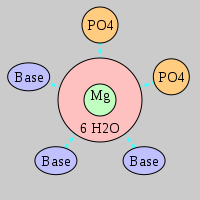
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4ton |
A1622 |
2Pout·3Bout |
|
|
|
A1800, A1801, A1802, A1803, A1804, A1805 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
4ton |
A1608 |
2Pout·3Bout |
|
|
|
A1732, A1733, A1734, A1735, A1736, A1737 |
|
A:A315, A:A329 |
|
G:A319, A:A320, G:A318 |
|
4tov |
A3019 |
2Pout·3Bout |
|
|
|
A3287, A3288, A3289, A3290, A3291, E303 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
4tov |
A3094 |
2Pout·3Bout |
|
|
|
A3628, A3629, A3630, A3631, A3632, A3633 |
|
A:A1572, G:A1422 |
|
G:A1423, G:A1424, G:A1425 |
|
4tow |
A1622 |
2Pout·3Bout |
|
|
|
A1799, A1800, A1801, A1802, A1803, A1804 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
4tow |
A1606 |
2Pout·3Bout |
|
|
|
A1724, A1725, A1726, A1727, T101, T102 |
|
G:A258, G:A259 |
|
G:A258, G:A259, G:A260 |
|
4tow |
A1608 |
2Pout·3Bout |
|
|
|
A1732, A1733, A1734, A1735, A1736, A1737 |
|
A:A315, A:A329 |
|
G:A319, A:A320, G:A318 |
|
4tp1 |
A3094 |
2Pout·3Bout |
|
|
|
A3628, A3629, A3630, A3631, A3632, A3633 |
|
A:A1572, G:A1422 |
|
G:A1423, G:A1424, G:A1425 |
|
4tp1 |
A3019 |
2Pout·3Bout |
|
|
|
A3287, A3288, A3289, A3290, A3291, A3292 |
|
A:A616, A:A608 |
|
G:A618, G:A619, G:A617 |
|
4tp1 |
A3133 |
2Pout·3Bout |
|
|
|
A3787, A3788, C405, C406, C407 |
|
C:A2591, G:A2592 |
|
G:A2592, U:A2593, A:A2600 |
|
4tp2 |
A1606 |
2Pout·3Bout |
|
|
|
A1724, A1725, A1726, A1727, A1728, T101 |
|
G:A258, G:A259 |
|
G:A258, G:A260, G:A259 |
|
4tp3 |
A3038 |
2Pout·3Bout |
|
|
|
A3372, A3373, A3374, A3375, A3376, A3377 |
|
U:A1267, A:A2009 |
|
A:A2009, G:A2010, U:A2011 |
|
4tp5 |
A3019 |
2Pout·3Bout |
|
|
|
A3287, A3288, A3289, A3290, E303, E304 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
4tp6 |
A1608 |
2Pout·3Bout |
|
|
|
A1732, A1733, A1734, A1735, A1736, A1737 |
|
A:A315, A:A329 |
|
G:A319, A:A320, G:A318 |
|
4tp6 |
A1622 |
2Pout·3Bout |
|
|
|
A1801, A1802, A1803, A1804, A1805, A1806 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
4tpa |
A1606 |
2Pout·3Bout |
|
|
|
A1724, A1725, A1726, A1727, A1728, T101 |
|
G:A258, G:A259 |
|
G:A258, G:A260, G:A259 |
|
4tpb |
A3085 |
2Pout·3Bout |
|
|
|
A3582, A3583, A3584, A3585, A3586, A3587 |
|
G:A977, G:A976 |
|
G:A977, G:A978, A:A979 |
|
4tpb |
A3038 |
2Pout·3Bout |
|
|
|
A3372, A3373, A3374, A3375, A3376, A3377 |
|
U:A1267, A:A2009 |
|
A:A2009, G:A2010, U:A2011 |
|
4tpc |
A1611 |
2Pout·3Bout |
|
|
|
A1750, A1751, A1752, A1753, A1754, A1755 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
4tpd |
A3093 |
2Pout·3Bout |
|
|
|
A3630, A3631, A3632, A3633, A3634, A3635 |
|
A:A1572, G:A1422 |
|
G:A1423, G:A1424, G:A1425 |
|
4tpd |
A3019 |
2Pout·3Bout |
|
|
|
A3288, A3289, A3290, A3291, E303, E304 |
|
A:A616, A:A608 |
|
G:A617, G:A618, G:A619 |
|
4tpe |
A1622 |
2Pout·3Bout |
|
|
|
A1802, A1803, A1804, A1805, A1806, A1807 |
|
G:A803, U:A804 |
|
G:A771, U:A772, G:A773 |
|
4tpe |
A1608 |
2Pout·3Bout |
|
|
|
A1733, A1734, A1735, A1736, A1737, A1738 |
|
A:A315, A:A329 |
|
G:A319, A:A320, G:A318 |
|
4tpf |
A3019 |
2Pout·3Bout |
|
|
|
A3285, A3286, A3287, A3288, A3289, E305 |
|
A:A616, G:A617 |
|
G:A617, G:A618, G:A619 |
|