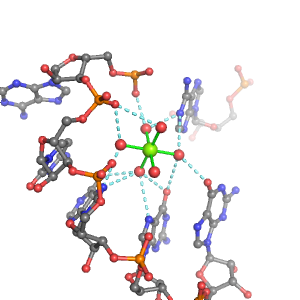
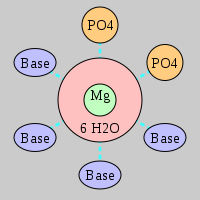
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1vs6 |
B2922 |
2Pout·4Bout |
|
|
|
B3098, B3099, B3100, B3101, B3102, B3103 |
|
A:B2009, U:B1267 |
|
G:B2010, U:B2011, G:B2012, A:B2009 |
|
1vs8 |
B2930 |
2Pout·4Bout |
|
|
|
B3139, B3140, B3141, B3142, B3143, B3144 |
|
C:B2452, A:B2453 |
|
A:B2453, G:B2454, A:B2497, G:B2455 |
|
1vt2 |
A3003 |
2Pout·4Bout |
|
|
|
A3509, A3510, A3511, A3512, A3513 |
|
A:A2705, C:A2704 |
|
A:A2705, A:A2706, U:A2701, U:A2707 |
|
2i2t |
B3016 |
2Pout·4Bout |
|
|
|
B3523, B3524, B3525, B3526, C278, C279 |
|
C:B1790, A:B1791 |
|
G:B1826, U:B1827, G:B1792, G:B1828 |
|
2otj |
08071 |
2Pout·4Bout |
|
|
|
04142, 06085, 07434, 07562, 07831, 07832 |
|
A:01815, C:01816 |
|
A:01759, U:01813, C:01818, U:01817 |
|
2qal |
A2031 |
2Pout·4Bout |
|
|
|
A2032, A2033, A2034, A2035, A2036, A2037 |
|
G:A645, U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
2qam |
B3457 |
2Pout·4Bout |
|
|
|
B3985, B3986, B3987, B3988, B3989, B3990 |
|
A:B2451, C:B2452 |
|
A:B2497, G:B2455, A:B2453, G:B2454 |
|
2qao |
B3152 |
2Pout·4Bout |
|
|
|
B3743, B3744, B3745, B3746, B3747, B3748 |
|
A:B2453, C:B2452 |
|
A:B2453, G:B2454, A:B2497, G:B2455 |
|
2qb9 |
A2031 |
2Pout·4Bout |
|
|
|
A2384, A2385, A2386, A2387, A2388, A2389 |
|
G:A645, U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
2qba |
B3457 |
2Pout·4Bout |
|
|
|
B3982, B3983, B3984, B3985, B3986, B3987 |
|
A:B2451, C:B2452 |
|
A:B2453, G:B2454, A:B2497, G:B2455 |
|
2qbc |
B3152 |
2Pout·4Bout |
|
|
|
B3745, B3746, B3747, B3748, B3749, B3750 |
|
C:B2452, A:B2453 |
|
A:B2453, G:B2454, A:B2497, G:B2455 |
|
2qbe |
B3457 |
2Pout·4Bout |
|
|
|
B3982, B3983, B3984, B3985, B3986, B3987 |
|
A:B2451, C:B2452 |
|
A:B2497, G:B2455, A:B2453, G:B2454 |
|
2qbh |
A1548 |
2Pout·4Bout |
|
|
|
A1629, A1630, A1631, A1632, A1633, A1634 |
|
G:A645, U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
2qbk |
B2930 |
2Pout·4Bout |
|
|
|
B3141, B3142, B3143, B3144, B3145, B3146 |
|
A:B2453, C:B2452 |
|
A:B2453, G:B2454, A:B2497, G:B2455 |
|
2qou |
A1548 |
2Pout·4Bout |
|
|
|
A1922, A1923, A1924, A1925, A1926, A1927 |
|
G:A645, U:A644 |
|
G:A645, U:A593, U:A644, G:A646 |
|
3cc2 |
08071 |
2Pout·4Bout |
|
|
|
03670, 05625, 06994, 07126, 07398, 07399 |
|
A:01815, C:01816 |
|
A:01759, U:01813, C:01818, U:01817 |
|
3i1p |
A2980 |
2Pout·4Bout |
|
|
|
A3397, A3398, A3399, A3400, A3401, A3402 |
|
G:A2454, A:A2453 |
|
G:A2455, C:A2496, G:A2454, A:A2453 |
|
3i1t |
A2978 |
2Pout·4Bout |
|
|
|
A3389, A3390, A3391, A3392, A3393, A3394 |
|
U:A2387, A:A2386 |
|
C:A2326, A:A2386, G:A2389, U:A2387 |
|
3i20 |
A2974 |
2Pout·4Bout |
|
|
|
A3369, A3370, A3371, A3372, A3373, A3374 |
|
A:A2453, C:A2452 |
|
A:A2497, G:A2454, G:A2455, A:A2453 |
|
3i21 |
A1553 |
2Pout·4Bout |
|
|
|
A1681, A1682, A1683, A1684, A1685, A1686 |
|
C:A797, A:A781 |
|
U:A798, G:A799, G:A800, C:A797 |
|
3i22 |
A2919 |
2Pout·4Bout |
|
|
|
A3115, A3116, A3117, A3118, A3119, A3120 |
|
C:A1257, G:A579 |
|
U:A580, C:A581, U:A1258, G:A1259 |
|
3oas |
A2979 |
2Pout·4Bout |
|
|
|
A3398, A3399, A3400, A3401, A3402, A3403 |
|
G:A2454, A:A2453 |
|
G:A2455, C:A2496, G:A2454, A:A2453 |
|
3oat |
A2905 |
2Pout·4Bout |
|
|
|
A3044, A3045, A3046, A3047, A3048, A3049 |
|
U:A29, A:A447 |
|
G:A473, G:A474, U:A29, G:A30 |
|
3ofc |
A2905 |
2Pout·4Bout |
|
|
|
A3045, A3046, A3047, A3048, A3049, A3050 |
|
U:A29, A:A447 |
|
G:A473, G:A474, U:A29, G:A30 |
|
3ofq |
A3003 |
2Pout·4Bout |
|
|
|
A3504, A3505, A3506, A3507, A3508 |
|
A:A2705, C:A2704 |
|
A:A2705, A:A2706, U:A2701, U:A2707 |
|