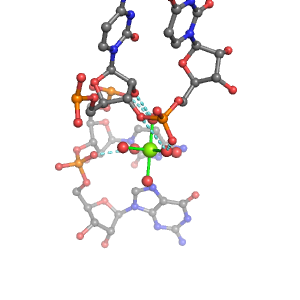
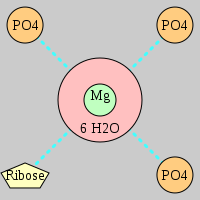
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2z4k |
A1566 |
3Pout·Rout |
|
|
|
A1712, A1713, A1714, A1715, A1716, A1717 |
|
U:A854, G:A727, C:A726 |
C:A853 |
|
|
3i1o |
A1570 |
3Pout·Rout |
|
|
|
A1761, A1762, A1763, A1764, A1765 |
|
A:A523, C:A536, C:A503 |
C:A522 |
|
|
3ofx |
A1573 |
3Pout·Rout |
|
|
|
A1763, A1764, A1765, A1766, A1767, A1768 |
|
C:A504, C:A536, C:A503 |
C:A522 |
|
|
3ofa |
A1573 |
3Pout·Rout |
|
|
|
A1763, A1764, A1765, A1766, A1767, A1768 |
|
C:A504, C:A503, C:A536 |
C:A522 |
|
|
3i1m |
A1574 |
3Pout·Rout |
|
|
|
A1764, A1765, A1766, A1767, A1768, L240 |
|
C:A504, C:A536, C:A503 |
C:A522 |
|
|
3oaq |
A1574 |
3Pout·Rout |
|
|
|
A1764, A1765, A1766, A1767, A1768, A1769 |
|
C:A504, C:A503, C:A536 |
C:A522 |
|
|
3i1z |
A1574 |
3Pout·Rout |
|
|
|
A1762, A1763, A1764, A1765, A1766, L240 |
|
C:A504, C:A536, C:A503 |
C:A522 |
|
|
3i1q |
A1574 |
3Pout·Rout |
|
|
|
A1759, A1760, A1761, A1762, A1763, L240 |
|
C:A504, C:A536, C:A503 |
C:A522 |
|
|
3or9 |
A1574 |
3Pout·Rout |
|
|
|
A1763, A1764, A1765, A1766, A1767, A1768 |
|
C:A504, C:A503, C:A536 |
C:A522 |
|
|
2qbh |
A1589 |
3Pout·Rout |
|
|
|
A1823, A1824, A1825, A1826, A1827, A1828 |
|
C:A25, U:A552, A:A553 |
G:A524 |
|
|
4gas |
A1615 |
3Pout·Rout |
|
|
|
A1768, A1769, A1770, A1771, A1772, A1773 |
|
C:A536, G:A537, C:A504 |
C:A522 |
|
|
4ton |
A1615 |
3Pout·Rout |
|
|
|
A1765, A1766, A1767, A1768, A1769, A1770 |
|
C:A504, C:A503, C:A536 |
C:A522 |
|
|
4duz |
A1628 |
3Pout·Rout |
|
|
|
A1956, A1957, A1958, A1959 |
|
A:A563, U:A560, C:A562 |
A:A559 |
|
|
4gaq |
A1629 |
3Pout·Rout |
|
|
|
A1836, A1837, A1838, A1839 |
|
G:A1198, G:A1053, C:A1054 |
G:A1198 |
|
|
4gd2 |
A1631 |
3Pout·Rout |
|
|
|
A1846, A1847, A1848, A1849, N204, N205 |
|
U:A1049, G:A1048, A:A1204 |
C:A1203 |
|
|
4dr2 |
A1676 |
3Pout·Rout |
|
|
|
A2167, A2168, A2169, L302 |
|
C:A536, G:A537, A:A523 |
C:A522 |
|
|
3v26 |
A1721 |
3Pout·Rout |
|
|
|
A1994, A1995, A1996, A1997, A1998, A1999 |
|
G:A406, A:A432, A:A496 |
A:A495 |
|
|
4dv0 |
A1787 |
3Pout·Rout |
|
|
|
A2134, A2135, A2136, A2137, A2138 |
|
C:A150, A:A151, G:A168 |
G:A167 |
|
|
4duz |
A1810 |
3Pout·Rout |
|
|
|
A2102, A2103, A2104, A2105, A2106 |
|
C:A150, A:A151, G:A168 |
G:A167 |
|
|
4dv0 |
A1823 |
3Pout·Rout |
|
|
|
A2260, A2261, A2262, A2263, A2264 |
|
C:A48, U:A49, G:A115 |
C:A47 |
|
|
4dr5 |
A1824 |
3Pout·Rout |
|
|
|
A2334, A2335, A2336, A2337, A2338, A2339 |
|
C:A904, U:A891, G:A903 |
G:A890 |
|
|
4dv6 |
A1853 |
3Pout·Rout |
|
|
|
A2344, A2345, A2346, A2347, A2348 |
|
C:A328, C:A330, C:A314 |
A:A329 |
|
|
4ji7 |
A1976 |
3Pout·Rout |
|
|
|
A3360, A3361, A3362, A3363, A3364, L307 |
|
C:A536, G:A537, C:A503 |
C:A522 |
|
|
2qal |
A2136 |
3Pout·Rout |
|
|
|
A2137, A2138, A2139, A2140, A2141, A2142 |
|
U:A854, G:A727, C:A726 |
C:A853 |
|
|
1vs6 |
B2908 |
3Pout·Rout |
|
|
|
B3031, B3032, B3033, B3034, B3035, B3036 |
|
C:B560, A:B1008, G:B559 |
C:B1007 |
|
|