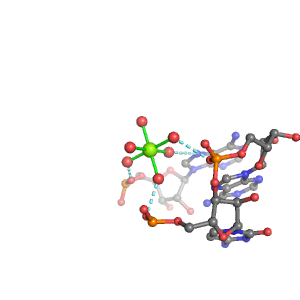
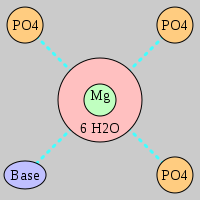
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3ud3 |
R1 |
3Pout·Bout |
|
|
|
R671, R672, R673, R674, R675, R676 |
|
G:R40, G:R41, A:R23 |
|
G:R42 |
|
2z4m |
E167 |
3Pout·Bout |
|
|
|
A1727, A1728, A1729, A1730, E171 |
|
U:A20, G:A21, U:A561 |
|
G:A21 |
|
3v27 |
B218 |
3Pout·Bout |
|
|
|
B330, B331, B332, B333, B334 |
|
U:B74, A:B100, G:B75 |
|
G:B76 |
|
4dha |
B221 |
3Pout·Bout |
|
|
|
B314, B315, B316, B317 |
|
A:B102, G:B103, A:B73 |
|
G:B103 |
|
2qow |
E291 |
3Pout·Bout |
|
|
|
A2117, A2118, A2119, E295, E297, E1832 |
|
U:A561, U:A20, G:A21 |
|
G:A21 |
|
2z4k |
A1551 |
3Pout·Bout |
|
|
|
A1640, A1641, A1642, A1643, A1644, A1645 |
|
A:A315, U:A317, C:A316 |
|
G:A318 |
|
3i1z |
A1552 |
3Pout·Bout |
|
|
|
A1663, A1664, A1665, A1666, A1667, E167 |
|
G:A1077, G:A1079, U:A1076 |
|
G:A1077 |
|
2qou |
A1559 |
3Pout·Bout |
|
|
|
A1974, A1975, A1976, A1977, A1978, A1979 |
|
A:A16, U:A13, U:A14 |
|
U:A14 |
|
2qou |
A1569 |
3Pout·Bout |
|
|
|
A2029, A2030, A2031, A2032, A2033 |
|
A:A767, A:A768, U:A804 |
|
G:A769 |
|
2i2p |
A1569 |
3Pout·Bout |
|
|
|
A1736, A1737, A1738, A1739, A1740 |
|
A:A768, A:A767, U:A804 |
|
G:A769 |
|
2qbh |
A1569 |
3Pout·Bout |
|
|
|
A1731, A1732, A1733, A1734, A1735 |
|
A:A768, U:A804, A:A767 |
|
G:A769 |
|
2aw7 |
A1573 |
3Pout·Bout |
|
|
|
A1759, A1760, A1761, A1762, A1763, A1764 |
|
A:A768, C:A805, A:A767 |
|
G:A769 |
|
1vs7 |
A1574 |
3Pout·Bout |
|
|
|
A1757, A1758, A1759, A1760, A1761, A1762 |
|
A:A768, C:A805, A:A767 |
|
G:A769 |
|
2avy |
A1597 |
3Pout·Bout |
|
|
|
A1862, A1863, A1864, A1865, A1866, A1867 |
|
A:A919, A:A1394, A:A918 |
|
A:A919 |
|
2qoy |
A1598 |
3Pout·Bout |
|
|
|
A2164, A2165, A2166, A2167, A2168, A2169 |
|
A:A1394, G:A917, A:A918 |
|
A:A919 |
|
1vs5 |
A1598 |
3Pout·Bout |
|
|
|
A1866, A1867, A1868, A1869, A1870, A1871 |
|
A:A919, A:A1394, A:A918 |
|
A:A919 |
|
2z4k |
A1599 |
3Pout·Bout |
|
|
|
A1869, A1870, A1871, A1872, A1873, A1874 |
|
A:A1394, G:A917, A:A918 |
|
A:A919 |
|
4ji1 |
A1606 |
3Pout·Bout |
|
|
|
A1912, A1913, A1914, A1915, A1916 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4gaq |
A1609 |
3Pout·Bout |
|
|
|
A1733, A1734, A1735, A1736, A1737, A1738 |
|
A:A315, G:A319, A:A329 |
|
G:A319 |
|
4dv7 |
A1611 |
3Pout·Bout |
|
|
|
A1923, A1924, A1925, A1926, A1927 |
|
G:A617, C:A620, A:A621 |
|
G:A617 |
|
4duy |
A1611 |
3Pout·Bout |
|
|
|
A1927, A1928, A1929, A1930, A1931 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4dv5 |
A1611 |
3Pout·Bout |
|
|
|
A1923, A1924, A1925, A1926, A1927 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4duz |
A1611 |
3Pout·Bout |
|
|
|
A1923, A1924, A1925, A1926, A1927 |
|
G:A617, C:A620, A:A621 |
|
G:A617 |
|
4dv6 |
A1611 |
3Pout·Bout |
|
|
|
A1927, A1928, A1929, A1930, D401 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4dv4 |
A1612 |
3Pout·Bout |
|
|
|
A1926, A1927, A1928, A1929, A1930 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|