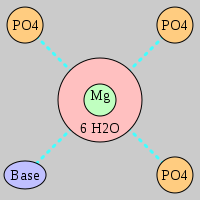
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4bpo |
A5030 |
3Pout·Bout |
|
|
|
A2043, A2044, A2045, A2046, A2047, A2379 |
|
U:A140, A:A141, G:A159 |
|
G:A159 |
|
4bpp |
A5030 |
3Pout·Bout |
|
|
|
A7491, A7492, A7493, A7494, A7495, A7496 |
|
U:A140, G:A159, A:A141 |
|
G:A159 |
|
4dha |
A3663 |
3Pout·Bout |
|
|
|
A4603, A4604, A4605, A4606 |
|
G:A2578, C:A2050, A:A2614 |
|
A:A2614 |
|
4dha |
B221 |
3Pout·Bout |
|
|
|
B314, B315, B316, B317 |
|
A:B102, G:B103, A:B73 |
|
G:B103 |
|
4dha |
A3703 |
3Pout·Bout |
|
|
|
A4747, A4748, A4749, A4750 |
|
A:A1986, C:A2551, A:A1669 |
|
G:A1954 |
|
4dhc |
A3685 |
3Pout·Bout |
|
|
|
A4705, A4706, A4707, A4708, A4709, A4710 |
|
A:A2657, C:A2658, G:A2659 |
|
G:A2659 |
|
4dhc |
A3630 |
3Pout·Bout |
|
|
|
A4499, A4500, A4501, A4502, A4503 |
|
G:A1987, C:A2551, A:A1669 |
|
G:A1954 |
|
4dhc |
A3447 |
3Pout·Bout |
|
|
|
A4015, A4016, A4017, A4018, A4019 |
|
U:A475, A:A478, G:A476 |
|
G:A476 |
|
4dr1 |
A1613 |
3Pout·Bout |
|
|
|
A1927, A1928, A1929, A1930, A1931 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4dr2 |
A1625 |
3Pout·Bout |
|
|
|
A2024, A2025, A2026, A2027 |
|
C:A651, U:A751, G:A752 |
|
G:A752 |
|
4dr3 |
A1613 |
3Pout·Bout |
|
|
|
A1925, A1926, A1927, A1928, A1929 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4dr3 |
A1827 |
3Pout·Bout |
|
|
|
A2142, A2143, A2144, A2145, A2146, A2147 |
|
C:A1103, A:A1101, C:A1100 |
|
C:A1100 |
|
4dr4 |
A1761 |
3Pout·Bout |
|
|
|
A2327, A2328, A2329, A2330 |
|
C:A458, G:A1300, C:A461 |
|
G:A459 |
|
4dr5 |
A1811 |
3Pout·Bout |
|
|
|
A2256, A2257, A2258, A2259, A2260, A2261 |
|
C:A150, G:A168, G:A167 |
|
G:A168 |
|
4dr6 |
A1612 |
3Pout·Bout |
|
|
|
A2033, A2034, A2035, A2036, A2037 |
|
C:A620, G:A617, A:A621 |
|
G:A617 |
|
4duy |
A1611 |
3Pout·Bout |
|
|
|
A1927, A1928, A1929, A1930, A1931 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4duz |
A1611 |
3Pout·Bout |
|
|
|
A1923, A1924, A1925, A1926, A1927 |
|
G:A617, C:A620, A:A621 |
|
G:A617 |
|
4dv0 |
A1807 |
3Pout·Bout |
|
|
|
A2203, A2204, A2205, A2206, A2207, A2208 |
|
A:A171, G:A148, A:A149 |
|
G:A148 |
|
4dv1 |
A1767 |
3Pout·Bout |
|
|
|
A2106, A2107, A2108, A2109, A2110 |
|
C:A1103, C:A1100, A:A1101 |
|
C:A1100 |
|
4dv3 |
A1612 |
3Pout·Bout |
|
|
|
A1923, A1924, A1925, A1926, A1927 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4dv4 |
A1612 |
3Pout·Bout |
|
|
|
A1926, A1927, A1928, A1929, A1930 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4dv5 |
A1776 |
3Pout·Bout |
|
|
|
A2095, A2096, A2097, A2098, A2099, B401 |
|
C:A1103, C:A1100, A:A1101 |
|
C:A1100 |
|
4dv5 |
A1611 |
3Pout·Bout |
|
|
|
A1923, A1924, A1925, A1926, A1927 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4dv6 |
A1611 |
3Pout·Bout |
|
|
|
A1927, A1928, A1929, A1930, D401 |
|
C:A620, A:A621, G:A617 |
|
G:A617 |
|
4dv7 |
A1611 |
3Pout·Bout |
|
|
|
A1923, A1924, A1925, A1926, A1927 |
|
G:A617, C:A620, A:A621 |
|
G:A617 |
|