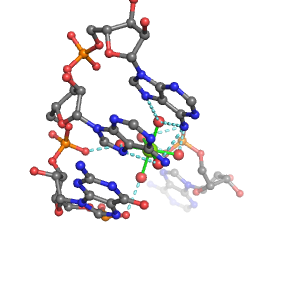
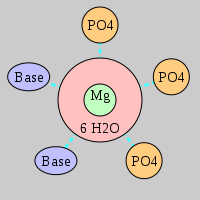
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3i21 |
A2 |
3Pout·2Bout |
|
|
|
A1577, A1578, A1579, A1580, A1581, A1582 |
|
A:A315, A:A329, G:A319 |
|
A:A320, G:A319 |
|
1l2x |
A38 |
3Pout·2Bout |
|
|
|
A39, A40, A41, A42, A43, A44 |
|
C:A26, G:A27, G:A28 |
|
G:A27, G:A28 |
|
3owz |
A115 |
3Pout·2Bout |
|
|
|
A141, A142, A143, A144, A145, A146 |
|
U:A23, C:A24, G:A25 |
|
G:A25, G:A26 |
|
3v23 |
B211 |
3Pout·2Bout |
|
|
|
B313, B314, B315, B316, B317 |
|
U:B74, A:B100, G:B75 |
|
G:B75, G:B76 |
|
4gaq |
D301 |
3Pout·2Bout |
|
|
|
A1774, A1775, A1776, D401, D402 |
ARG:D70(NH1) |
C:A401, A:A547, G:A548 |
|
G:A402, C:A401 |
|
1dfu |
N504 |
3Pout·2Bout |
|
|
|
M536, N507, N527, N528, N530, N568 |
|
A:M99, G:N75, U:N74 |
|
G:N76, G:N75 |
|
2qoy |
A1550 |
3Pout·2Bout |
|
|
|
A1935, A1936, A1937, A1938, A1939, A1940 |
|
A:A315, A:A329, C:A316 |
|
G:A318, G:A319 |
|
2qp0 |
A1560 |
3Pout·2Bout |
|
|
|
A1975, A1976, A1977, A1978, A1979, A1980 |
|
A:A768, C:A805, A:A767 |
|
A:A768, G:A769 |
|
2qbj |
A1572 |
3Pout·2Bout |
|
|
|
A1755, A1756, A1757, A1758, A1759, A1760 |
|
A:A768, C:A805, A:A767 |
|
A:A768, G:A769 |
|
2i2u |
A1574 |
3Pout·2Bout |
|
|
|
A1745, A1746, A1747, A1748, A1749, A1750 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
2i2u |
A1596 |
3Pout·2Bout |
|
|
|
A1856, A1857, A1858, A1859, X267, X270 |
|
G:A1401, U:X2, G:X3 |
|
C:A1399, G:A1401 |
|
2qbf |
A1600 |
3Pout·2Bout |
|
|
|
A1886, A1887, A1888, A1889, A1890, A1891 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
2qbj |
A1600 |
3Pout·2Bout |
|
|
|
A1883, A1884, A1885, A1886, A1887, A1888 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
2aw7 |
A1601 |
3Pout·2Bout |
|
|
|
A1888, A1889, A1890, A1891, A1892, A1893 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
1vs7 |
A1602 |
3Pout·2Bout |
|
|
|
A1886, A1887, A1888, A1889, A1890, A1891 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
4ji6 |
A1607 |
3Pout·2Bout |
|
|
|
A2020, A2021, A2022, A2023, A2024 |
|
G:A617, A:A621, C:A620 |
|
G:A616, G:A617 |
|
4ji2 |
A1613 |
3Pout·2Bout |
|
|
|
A1927, A1928, A1929, A1930, A1931 |
|
C:A620, A:A621, G:A617 |
|
G:A617, G:A616 |
|
3v2c |
A1673 |
3Pout·2Bout |
|
|
|
A2018, A2019, A2020, A2021, A2022 |
|
A:A780, G:A799, C:A779 |
|
G:A800, A:A780 |
|
4ji5 |
A1718 |
3Pout·2Bout |
|
|
|
A2033, A2034, A2035, A2036, A2037, A2038 |
|
G:A869, G:A858, C:A868 |
|
G:A858, G:A869 |
|
3v26 |
A1722 |
3Pout·2Bout |
|
|
|
A2000, A2001, A2002, A2003 |
|
A:A780, G:A799, G:A800 |
|
G:A800, A:A780 |
|
4dv0 |
A1788 |
3Pout·2Bout |
|
|
|
A2139, A2140, A2141, A2142 |
|
C:A307, C:A308, G:A306 |
|
G:A309, G:A310 |
|
4dv0 |
A1799 |
3Pout·2Bout |
|
|
|
A2174, A2175, A2176, A2177, A2178, A2179 |
|
C:A857, C:A868, G:A869 |
|
G:A858, G:A869 |
|
4dv4 |
A1800 |
3Pout·2Bout |
|
|
|
A2163, A2164, A2165, A2166, A2167, A2168 |
|
C:A868, G:A869, C:A857 |
|
G:A858, G:A869 |
|
4dv1 |
A1820 |
3Pout·2Bout |
|
|
|
A2232, A2233, A2234, A2235, A2236, A2237 |
|
U:A757, U:A580, G:A758 |
|
G:A581, G:A758 |
|
4dv1 |
A1821 |
3Pout·2Bout |
|
|
|
A2238, A2239, A2240, A2241, A2242, A2243 |
|
C:A868, G:A869, C:A857 |
|
G:A858, G:A869 |
|