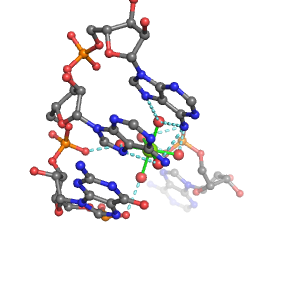
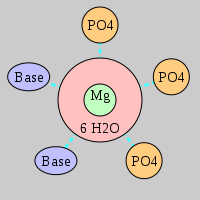
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3v29 |
A3382 |
3Pout·2Bout |
|
|
|
A3940, A3941, A3942, A3943, A3944, A3945 |
|
A:A1634, U:A1300, G:A1627 |
|
G:A1628, U:A1629 |
|
3v29 |
A3374 |
3Pout·2Bout |
|
|
|
A3908, A3909, A3910, A3911, A3912, P303 |
|
G:A831, G:A944, U:A568 |
|
A:A2448, A:A945 |
|
3v2c |
A1673 |
3Pout·2Bout |
|
|
|
A2018, A2019, A2020, A2021, A2022 |
|
A:A780, G:A799, C:A779 |
|
G:A800, A:A780 |
|
3v2d |
A3363 |
3Pout·2Bout |
|
|
|
A4701, A4702, A4703, A4704, A4705, A4706 |
|
G:A1332, C:A1333, U:A1313 |
|
U:A1335, G:A1334 |
|
3v2f |
A3319 |
3Pout·2Bout |
|
|
|
A4318, A4319, A4320, A4321, A4322 |
|
G:A1358, U:A1357, C:A1370 |
|
G:A1371, G:A1358 |
|
4bpp |
A5069 |
3Pout·2Bout |
|
|
|
A8115, A8116, A8117, A8118, A8119, A8120 |
|
C:A1428, C:A1149, G:A1148 |
|
A:A1174, G:A1172 |
|
4dha |
A3731 |
3Pout·2Bout |
|
|
|
A4833, A4834, A4835, A4836, A4837, A4838 |
|
G:A770, G:A686, G:A771 |
|
G:A771, G:A770 |
|
4dr4 |
A1948 |
3Pout·2Bout |
|
|
|
A2434, A2435, A2436, A2437, A2438 |
|
A:A608, A:A609, A:A607 |
|
A:A609, G:A610 |
|
4duz |
A1840 |
3Pout·2Bout |
|
|
|
A2218, A2219, A2220, A2221, A2222 |
|
G:A858, C:A868, C:A857 |
|
G:A858, G:A869 |
|
4duz |
A1839 |
3Pout·2Bout |
|
|
|
A2212, A2213, A2214, A2215, A2216, A2217 |
|
G:A581, U:A757, U:A580 |
|
G:A758, G:A581 |
|
4dv0 |
A1799 |
3Pout·2Bout |
|
|
|
A2174, A2175, A2176, A2177, A2178, A2179 |
|
C:A857, C:A868, G:A869 |
|
G:A858, G:A869 |
|
4dv0 |
A1788 |
3Pout·2Bout |
|
|
|
A2139, A2140, A2141, A2142 |
|
C:A307, C:A308, G:A306 |
|
G:A309, G:A310 |
|
4dv1 |
A1821 |
3Pout·2Bout |
|
|
|
A2238, A2239, A2240, A2241, A2242, A2243 |
|
C:A868, G:A869, C:A857 |
|
G:A858, G:A869 |
|
4dv1 |
A1820 |
3Pout·2Bout |
|
|
|
A2232, A2233, A2234, A2235, A2236, A2237 |
|
U:A757, U:A580, G:A758 |
|
G:A581, G:A758 |
|
4dv3 |
A1848 |
3Pout·2Bout |
|
|
|
A2215, A2216, A2217, A2218, A2219, A2220 |
|
U:A757, G:A581, U:A580 |
|
G:A581, G:A758 |
|
4dv4 |
A1800 |
3Pout·2Bout |
|
|
|
A2163, A2164, A2165, A2166, A2167, A2168 |
|
C:A868, G:A869, C:A857 |
|
G:A858, G:A869 |
|
4dv6 |
A1824 |
3Pout·2Bout |
|
|
|
A2237, A2238, A2239, A2240, A2241, A2242 |
|
G:A858, C:A868, G:A869 |
|
G:A858, G:A869 |
|
4dv7 |
A1863 |
3Pout·2Bout |
|
|
|
A2234, A2235, A2236, A2237, A2238, A2239 |
|
G:A858, C:A868, C:A857 |
|
G:A858, G:A869 |
|
4dv7 |
A1862 |
3Pout·2Bout |
|
|
|
A2228, A2229, A2230, A2231, A2232, A2233 |
|
G:A581, U:A580, G:A758 |
|
G:A758, G:A581 |
|
4gaq |
D301 |
3Pout·2Bout |
|
|
|
A1774, A1775, A1776, D401, D402 |
ARG:D70(NH1) |
C:A401, A:A547, G:A548 |
|
G:A402, C:A401 |
|
4gau |
A3059 |
3Pout·2Bout |
|
|
|
A3481, A3482, A3483, A3484, A3485, A3486 |
|
G:A2578, U:A2613, A:A2051 |
|
A:A2051, A:A2614 |
|
4gau |
A3042 |
3Pout·2Bout |
|
|
|
A3401, A3402, A3403, A3404 |
|
G:A1358, C:A1357, C:A1370 |
|
G:A1358, G:A1371 |
|
4ji2 |
A1613 |
3Pout·2Bout |
|
|
|
A1927, A1928, A1929, A1930, A1931 |
|
C:A620, A:A621, G:A617 |
|
G:A617, G:A616 |
|
4ji5 |
A1718 |
3Pout·2Bout |
|
|
|
A2033, A2034, A2035, A2036, A2037, A2038 |
|
G:A869, G:A858, C:A868 |
|
G:A858, G:A869 |
|
4ji6 |
A1607 |
3Pout·2Bout |
|
|
|
A2020, A2021, A2022, A2023, A2024 |
|
G:A617, A:A621, C:A620 |
|
G:A616, G:A617 |
|