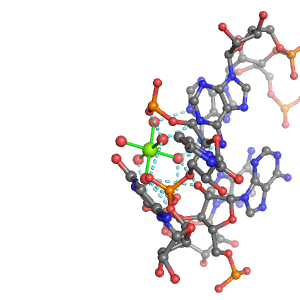
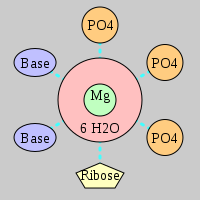
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3oxd |
A101 |
3Pout·Rout·2Bout |
|
|
|
A138, A139, A140, A141, A142, A143 |
|
C:A35, A:A65, A:A34 |
A:A81 |
G:A36, A:A37 |
|
3oxd |
B106 |
3Pout·Rout·2Bout |
|
|
|
A144, A145, B137, B138, B139, B140 |
|
C:A66, C:B66, A:B33 |
A:B65 |
A:B33, A:A65 |
|
3egz |
B501 |
3Pout·Rout·2Bout |
|
|
|
B301, B302, B303, B304, B305, B306 |
|
G:B10, G:B9, A:B22 |
A:B8 |
G:B10, U:B11 |
|
2i2p |
A1583 |
3Pout·Rout·2Bout |
|
|
|
A1800, A1801, A1802, A1803, A1804 |
|
A:A607, A:A608, G:A293 |
G:A292 |
C:A631, A:A630 |
|
4ji8 |
A1959 |
3Pout·Rout·2Bout |
|
|
|
A3258, A3259, A3260, A3261, A3262, A3263 |
|
C:A857, G:A869, C:A868 |
G:A858 |
G:A858, G:A869 |
|
3r8t |
A2950 |
3Pout·Rout·2Bout |
|
|
|
A3286, A3287, A3288, A3289, A3290 |
|
G:A1455, A:A1453, G:A1456 |
C:A1454 |
G:A1456, U:A1457 |
|
1vs8 |
B3010 |
3Pout·Rout·2Bout |
|
|
|
B3495, B3496, B3497, B3498, B3499, B3500 |
|
A:B973, G:B570, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2awb |
B3011 |
3Pout·Rout·2Bout |
|
|
|
B3496, B3497, B3498, B3499, B3500, B3501 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2qox |
B3011 |
3Pout·Rout·2Bout |
|
|
|
B3500, B3501, B3502, B3503, B3504, B3505 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2i2v |
B3011 |
3Pout·Rout·2Bout |
|
|
|
B3503, B3504, B3505, B3506, B3507, B3508 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2qbg |
B3012 |
3Pout·Rout·2Bout |
|
|
|
B3505, B3506, B3507, B3508, B3509, B3510 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2qov |
B3012 |
3Pout·Rout·2Bout |
|
|
|
B3500, B3501, B3502, B3503, B3504, B3505 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2qp1 |
B3012 |
3Pout·Rout·2Bout |
|
|
|
B4008, B4009, B4010, B4011, B4012, B4013 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
1vs6 |
B3012 |
3Pout·Rout·2Bout |
|
|
|
B3502, B3503, B3504, B3505, B3506, B3507 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2qoz |
B3012 |
3Pout·Rout·2Bout |
|
|
|
B3990, B3991, B3992, B3993, B3994, B3995 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2aw4 |
B3012 |
3Pout·Rout·2Bout |
|
|
|
B3498, B3499, B3500, B3501, B3502, B3503 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2i2t |
B3012 |
3Pout·Rout·2Bout |
|
|
|
B3506, B3507, B3508, B3509, B3510, B3511 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2qbk |
B3012 |
3Pout·Rout·2Bout |
|
|
|
B3498, B3499, B3500, B3501, B3502, B3503 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2z4l |
B3013 |
3Pout·Rout·2Bout |
|
|
|
B3498, B3499, B3500, B3501, B3502, B3503 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
2z4n |
B3013 |
3Pout·Rout·2Bout |
|
|
|
B3503, B3504, B3505, B3506, B3507, B3508 |
|
G:B570, A:B973, U:B569 |
A:B972 |
A:B820, A:B821 |
|
4tpb |
A3018 |
3Pout·Rout·2Bout |
|
|
|
A3281, A3282, A3283, A3284, A3285, A3286 |
|
G:A583, A:A1253, A:A582 |
C:A1251 |
G:A583, A:A582 |
|
4tox |
A3018 |
3Pout·Rout·2Bout |
|
|
|
A3280, A3281, A3282, A3283, A3284, A3285 |
|
G:A583, A:A1253, A:A582 |
C:A1251 |
G:A583, C:A584 |
|
4tp3 |
A3018 |
3Pout·Rout·2Bout |
|
|
|
A3281, A3282, A3283, A3284, A3285, A3286 |
|
A:A582, A:A1253, G:A583 |
C:A1251 |
G:A583, C:A584 |
|
3ofd |
A3027 |
3Pout·Rout·2Bout |
|
|
|
A3594, A3595, A3596, A3597, A3598, A3599 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, U:A568 |
|
3oas |
A3027 |
3Pout·Rout·2Bout |
|
|
|
A3593, A3594, A3595, A3596, A3597, A3598 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, U:A568 |
|